Karim Lekadir
Debiasing Cardiac Imaging with Controlled Latent Diffusion Models
Mar 28, 2024



Abstract:The progress in deep learning solutions for disease diagnosis and prognosis based on cardiac magnetic resonance imaging is hindered by highly imbalanced and biased training data. To address this issue, we propose a method to alleviate imbalances inherent in datasets through the generation of synthetic data based on sensitive attributes such as sex, age, body mass index, and health condition. We adopt ControlNet based on a denoising diffusion probabilistic model to condition on text assembled from patient metadata and cardiac geometry derived from segmentation masks using a large-cohort study, specifically, the UK Biobank. We assess our method by evaluating the realism of the generated images using established quantitative metrics. Furthermore, we conduct a downstream classification task aimed at debiasing a classifier by rectifying imbalances within underrepresented groups through synthetically generated samples. Our experiments demonstrate the effectiveness of the proposed approach in mitigating dataset imbalances, such as the scarcity of younger patients or individuals with normal BMI level suffering from heart failure. This work represents a major step towards the adoption of synthetic data for the development of fair and generalizable models for medical classification tasks. Notably, we conduct all our experiments using a single, consumer-level GPU to highlight the feasibility of our approach within resource-constrained environments. Our code is available at https://github.com/faildeny/debiasing-cardiac-mri.
Towards Learning Contrast Kinetics with Multi-Condition Latent Diffusion Models
Mar 20, 2024



Abstract:Contrast agents in dynamic contrast enhanced magnetic resonance imaging allow to localize tumors and observe their contrast kinetics, which is essential for cancer characterization and respective treatment decision-making. However, contrast agent administration is not only associated with adverse health risks, but also restricted for patients during pregnancy, and for those with kidney malfunction, or other adverse reactions. With contrast uptake as key biomarker for lesion malignancy, cancer recurrence risk, and treatment response, it becomes pivotal to reduce the dependency on intravenous contrast agent administration. To this end, we propose a multi-conditional latent diffusion model capable of acquisition time-conditioned image synthesis of DCE-MRI temporal sequences. To evaluate medical image synthesis, we additionally propose and validate the Fr\'echet radiomics distance as an image quality measure based on biomarker variability between synthetic and real imaging data. Our results demonstrate our method's ability to generate realistic multi-sequence fat-saturated breast DCE-MRI and uncover the emerging potential of deep learning based contrast kinetics simulation. We publicly share our accessible codebase at https://github.com/RichardObi/ccnet.
Pre- to Post-Contrast Breast MRI Synthesis for Enhanced Tumour Segmentation
Nov 17, 2023Abstract:Despite its benefits for tumour detection and treatment, the administration of contrast agents in dynamic contrast-enhanced MRI (DCE-MRI) is associated with a range of issues, including their invasiveness, bioaccumulation, and a risk of nephrogenic systemic fibrosis. This study explores the feasibility of producing synthetic contrast enhancements by translating pre-contrast T1-weighted fat-saturated breast MRI to their corresponding first DCE-MRI sequence leveraging the capabilities of a generative adversarial network (GAN). Additionally, we introduce a Scaled Aggregate Measure (SAMe) designed for quantitatively evaluating the quality of synthetic data in a principled manner and serving as a basis for selecting the optimal generative model. We assess the generated DCE-MRI data using quantitative image quality metrics and apply them to the downstream task of 3D breast tumour segmentation. Our results highlight the potential of post-contrast DCE-MRI synthesis in enhancing the robustness of breast tumour segmentation models via data augmentation. Our code is available at https://github.com/RichardObi/pre_post_synthesis.
USLR: an open-source tool for unbiased and smooth longitudinal registration of brain MR
Nov 14, 2023Abstract:We present USLR, a computational framework for longitudinal registration of brain MRI scans to estimate nonlinear image trajectories that are smooth across time, unbiased to any timepoint, and robust to imaging artefacts. It operates on the Lie algebra parameterisation of spatial transforms (which is compatible with rigid transforms and stationary velocity fields for nonlinear deformation) and takes advantage of log-domain properties to solve the problem using Bayesian inference. USRL estimates rigid and nonlinear registrations that: (i) bring all timepoints to an unbiased subject-specific space; and (i) compute a smooth trajectory across the imaging time-series. We capitalise on learning-based registration algorithms and closed-form expressions for fast inference. A use-case Alzheimer's disease study is used to showcase the benefits of the pipeline in multiple fronts, such as time-consistent image segmentation to reduce intra-subject variability, subject-specific prediction or population analysis using tensor-based morphometry. We demonstrate that such approach improves upon cross-sectional methods in identifying group differences, which can be helpful in detecting more subtle atrophy levels or in reducing sample sizes in clinical trials. The code is publicly available in https://github.com/acasamitjana/uslr
Revisiting Skin Tone Fairness in Dermatological Lesion Classification
Aug 18, 2023Abstract:Addressing fairness in lesion classification from dermatological images is crucial due to variations in how skin diseases manifest across skin tones. However, the absence of skin tone labels in public datasets hinders building a fair classifier. To date, such skin tone labels have been estimated prior to fairness analysis in independent studies using the Individual Typology Angle (ITA). Briefly, ITA calculates an angle based on pixels extracted from skin images taking into account the lightness and yellow-blue tints. These angles are then categorised into skin tones that are subsequently used to analyse fairness in skin cancer classification. In this work, we review and compare four ITA-based approaches of skin tone classification on the ISIC18 dataset, a common benchmark for assessing skin cancer classification fairness in the literature. Our analyses reveal a high disagreement among previously published studies demonstrating the risks of ITA-based skin tone estimation methods. Moreover, we investigate the causes of such large discrepancy among these approaches and find that the lack of diversity in the ISIC18 dataset limits its use as a testbed for fairness analysis. Finally, we recommend further research on robust ITA estimation and diverse dataset acquisition with skin tone annotation to facilitate conclusive fairness assessments of artificial intelligence tools in dermatology. Our code is available at https://github.com/tkalbl/RevisitingSkinToneFairness.
Commentary on explainable artificial intelligence methods: SHAP and LIME
May 08, 2023Abstract:eXplainable artificial intelligence (XAI) methods have emerged to convert the black box of machine learning models into a more digestible form. These methods help to communicate how the model works with the aim of making machine learning models more transparent and increasing the trust of end-users into their output. SHapley Additive exPlanations (SHAP) and Local Interpretable Model Agnostic Explanation (LIME) are two widely used XAI methods particularly with tabular data. In this commentary piece, we discuss the way the explainability metrics of these two methods are generated and propose a framework for interpretation of their outputs, highlighting their weaknesses and strengths.
Fairness and bias correction in machine learning for depression prediction: results from four different study populations
Nov 10, 2022Abstract:A significant level of stigma and inequality exists in mental healthcare, especially in under-served populations, which spreads through collected data. When not properly accounted for, machine learning (ML) models learned from data can reinforce the structural biases already present in society. Here, we present a systematic study of bias in ML models designed to predict depression in four different case studies covering different countries and populations. We find that standard ML approaches show regularly biased behaviors. However, we show that standard mitigation techniques, and our own post-hoc method, can be effective in reducing the level of unfair bias. We provide practical recommendations to develop ML models for depression risk prediction with increased fairness and trust in the real world. No single best ML model for depression prediction provides equality of outcomes. This emphasizes the importance of analyzing fairness during model selection and transparent reporting about the impact of debiasing interventions.
medigan: A Python Library of Pretrained Generative Models for Enriched Data Access in Medical Imaging
Sep 28, 2022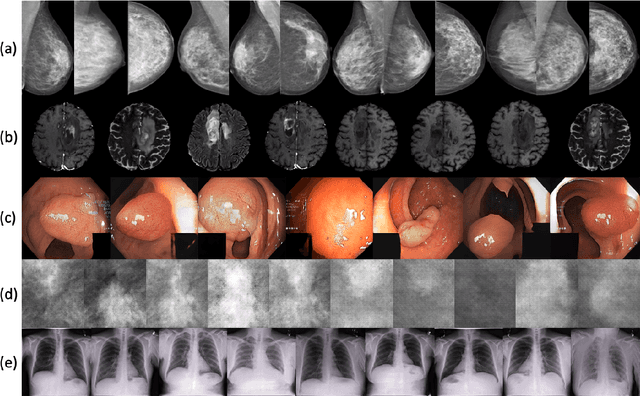
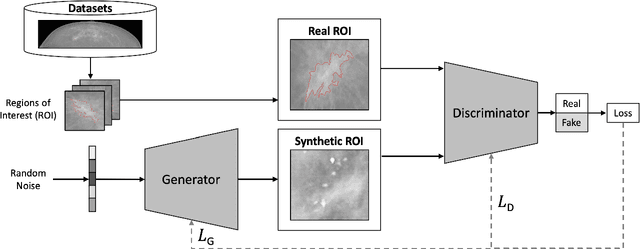
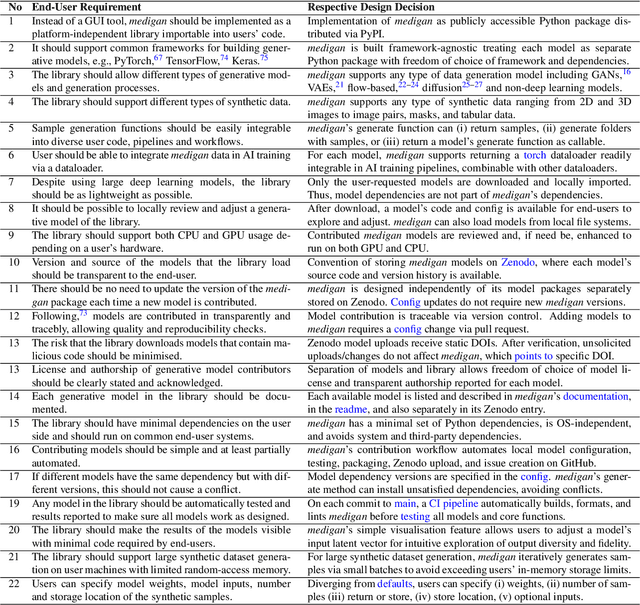
Abstract:Synthetic data generated by generative models can enhance the performance and capabilities of data-hungry deep learning models in medical imaging. However, there is (1) limited availability of (synthetic) datasets and (2) generative models are complex to train, which hinders their adoption in research and clinical applications. To reduce this entry barrier, we propose medigan, a one-stop shop for pretrained generative models implemented as an open-source framework-agnostic Python library. medigan allows researchers and developers to create, increase, and domain-adapt their training data in just a few lines of code. Guided by design decisions based on gathered end-user requirements, we implement medigan based on modular components for generative model (i) execution, (ii) visualisation, (iii) search & ranking, and (iv) contribution. The library's scalability and design is demonstrated by its growing number of integrated and readily-usable pretrained generative models consisting of 21 models utilising 9 different Generative Adversarial Network architectures trained on 11 datasets from 4 domains, namely, mammography, endoscopy, x-ray, and MRI. Furthermore, 3 applications of medigan are analysed in this work, which include (a) enabling community-wide sharing of restricted data, (b) investigating generative model evaluation metrics, and (c) improving clinical downstream tasks. In (b), extending on common medical image synthesis assessment and reporting standards, we show Fr\'echet Inception Distance variability based on image normalisation and radiology-specific feature extraction.
High-resolution synthesis of high-density breast mammograms: Application to improved fairness in deep learning based mass detection
Sep 20, 2022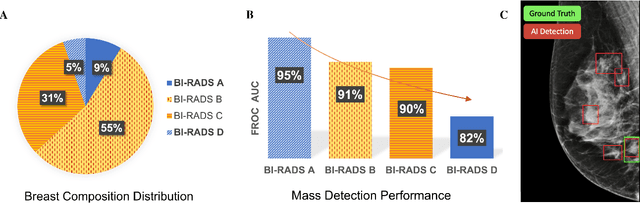
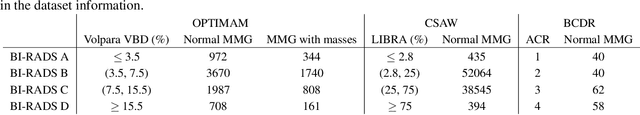
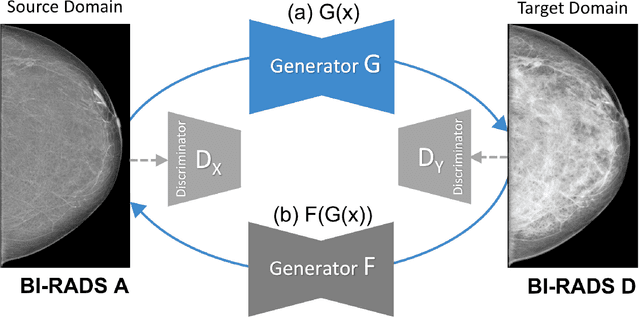
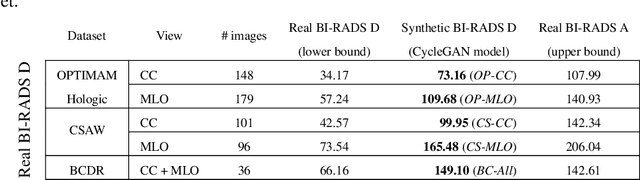
Abstract:Computer-aided detection systems based on deep learning have shown good performance in breast cancer detection. However, high-density breasts show poorer detection performance since dense tissues can mask or even simulate masses. Therefore, the sensitivity of mammography for breast cancer detection can be reduced by more than 20% in dense breasts. Additionally, extremely dense cases reported an increased risk of cancer compared to low-density breasts. This study aims to improve the mass detection performance in high-density breasts using synthetic high-density full-field digital mammograms (FFDM) as data augmentation during breast mass detection model training. To this end, a total of five cycle-consistent GAN (CycleGAN) models using three FFDM datasets were trained for low-to-high-density image translation in high-resolution mammograms. The training images were split by breast density BI-RADS categories, being BI-RADS A almost entirely fatty and BI-RADS D extremely dense breasts. Our results showed that the proposed data augmentation technique improved the sensitivity and precision of mass detection in high-density breasts by 2% and 6% in two different test sets and was useful as a domain adaptation technique. In addition, the clinical realism of the synthetic images was evaluated in a reader study involving two expert radiologists and one surgical oncologist.
Generalisability of deep learning models in low-resource imaging settings: A fetal ultrasound study in 5 African countries
Sep 20, 2022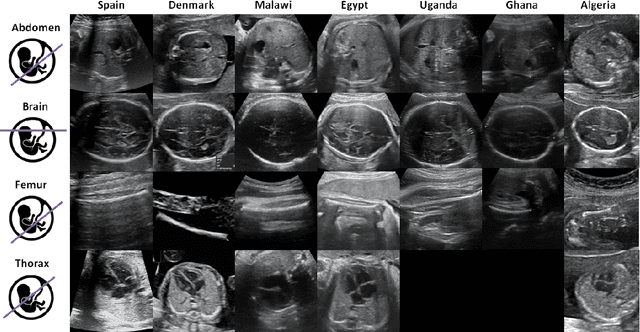
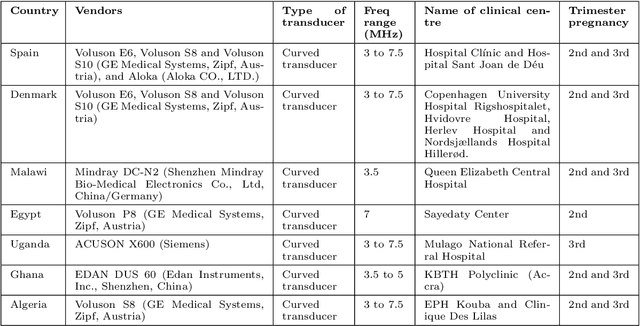
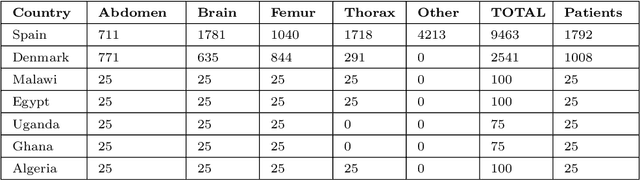
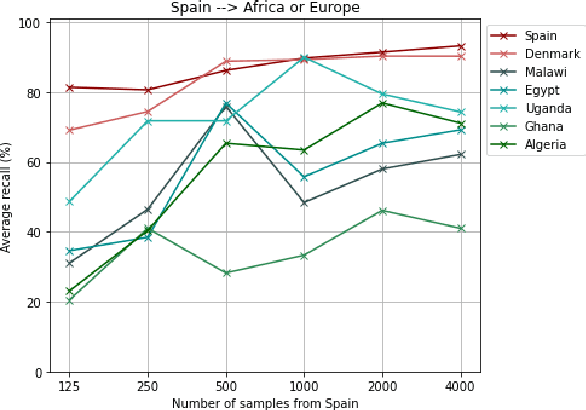
Abstract:Most artificial intelligence (AI) research have concentrated in high-income countries, where imaging data, IT infrastructures and clinical expertise are plentiful. However, slower progress has been made in limited-resource environments where medical imaging is needed. For example, in Sub-Saharan Africa the rate of perinatal mortality is very high due to limited access to antenatal screening. In these countries, AI models could be implemented to help clinicians acquire fetal ultrasound planes for diagnosis of fetal abnormalities. So far, deep learning models have been proposed to identify standard fetal planes, but there is no evidence of their ability to generalise in centres with limited access to high-end ultrasound equipment and data. This work investigates different strategies to reduce the domain-shift effect for a fetal plane classification model trained on a high-resource clinical centre and transferred to a new low-resource centre. To that end, a classifier trained with 1,792 patients from Spain is first evaluated on a new centre in Denmark in optimal conditions with 1,008 patients and is later optimised to reach the same performance in five African centres (Egypt, Algeria, Uganda, Ghana and Malawi) with 25 patients each. The results show that a transfer learning approach can be a solution to integrate small-size African samples with existing large-scale databases in developed countries. In particular, the model can be re-aligned and optimised to boost the performance on African populations by increasing the recall to $0.92 \pm 0.04$ and at the same time maintaining a high precision across centres. This framework shows promise for building new AI models generalisable across clinical centres with limited data acquired in challenging and heterogeneous conditions and calls for further research to develop new solutions for usability of AI in countries with less resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge