Fred Prior
Virtual-Eyes: Quantitative Validation of a Lung CT Quality-Control Pipeline for Foundation-Model Cancer Risk Prediction
Dec 30, 2025Abstract:Robust preprocessing is rarely quantified in deep-learning pipelines for low-dose CT (LDCT) lung cancer screening. We develop and validate Virtual-Eyes, a clinically motivated 16-bit CT quality-control pipeline, and measure its differential impact on generalist foundation models versus specialist models. Virtual-Eyes enforces strict 512x512 in-plane resolution, rejects short or non-diagnostic series, and extracts a contiguous lung block using Hounsfield-unit filtering and bilateral lung-coverage scoring while preserving the native 16-bit grid. Using 765 NLST patients (182 cancer, 583 non-cancer), we compute slice-level embeddings from RAD-DINO and Merlin with frozen encoders and train leakage-free patient-level MLP heads; we also evaluate Sybil and a 2D ResNet-18 baseline under Raw versus Virtual-Eyes inputs without backbone retraining. Virtual-Eyes improves RAD-DINO slice-level AUC from 0.576 to 0.610 and patient-level AUC from 0.646 to 0.683 (mean pooling) and from 0.619 to 0.735 (max pooling), with improved calibration (Brier score 0.188 to 0.112). In contrast, Sybil and ResNet-18 degrade under Virtual-Eyes (Sybil AUC 0.886 to 0.837; ResNet-18 AUC 0.571 to 0.596) with evidence of context dependence and shortcut learning, and Merlin shows limited transferability (AUC approximately 0.507 to 0.567) regardless of preprocessing. These results demonstrate that anatomically targeted QC can stabilize and improve generalist foundation-model workflows but may disrupt specialist models adapted to raw clinical context.
Medical Image De-Identification Benchmark Challenge
Jul 31, 2025



Abstract:The de-identification (deID) of protected health information (PHI) and personally identifiable information (PII) is a fundamental requirement for sharing medical images, particularly through public repositories, to ensure compliance with patient privacy laws. In addition, preservation of non-PHI metadata to inform and enable downstream development of imaging artificial intelligence (AI) is an important consideration in biomedical research. The goal of MIDI-B was to provide a standardized platform for benchmarking of DICOM image deID tools based on a set of rules conformant to the HIPAA Safe Harbor regulation, the DICOM Attribute Confidentiality Profiles, and best practices in preservation of research-critical metadata, as defined by The Cancer Imaging Archive (TCIA). The challenge employed a large, diverse, multi-center, and multi-modality set of real de-identified radiology images with synthetic PHI/PII inserted. The MIDI-B Challenge consisted of three phases: training, validation, and test. Eighty individuals registered for the challenge. In the training phase, we encouraged participants to tune their algorithms using their in-house or public data. The validation and test phases utilized the DICOM images containing synthetic identifiers (of 216 and 322 subjects, respectively). Ten teams successfully completed the test phase of the challenge. To measure success of a rule-based approach to image deID, scores were computed as the percentage of correct actions from the total number of required actions. The scores ranged from 97.91% to 99.93%. Participants employed a variety of open-source and proprietary tools with customized configurations, large language models, and optical character recognition (OCR). In this paper we provide a comprehensive report on the MIDI-B Challenge's design, implementation, results, and lessons learned.
MAMA-MIA: A Large-Scale Multi-Center Breast Cancer DCE-MRI Benchmark Dataset with Expert Segmentations
Jun 19, 2024Abstract:Current research in breast cancer Magnetic Resonance Imaging (MRI), especially with Artificial Intelligence (AI), faces challenges due to the lack of expert segmentations. To address this, we introduce the MAMA-MIA dataset, comprising 1506 multi-center dynamic contrast-enhanced MRI cases with expert segmentations of primary tumors and non-mass enhancement areas. These cases were sourced from four publicly available collections in The Cancer Imaging Archive (TCIA). Initially, we trained a deep learning model to automatically segment the cases, generating preliminary segmentations that significantly reduced expert segmentation time. Sixteen experts, averaging 9 years of experience in breast cancer, then corrected these segmentations, resulting in the final expert segmentations. Additionally, two radiologists conducted a visual inspection of the automatic segmentations to support future quality control studies. Alongside the expert segmentations, we provide 49 harmonized demographic and clinical variables and the pretrained weights of the well-known nnUNet architecture trained using the DCE-MRI full-images and expert segmentations. This dataset aims to accelerate the development and benchmarking of deep learning models and foster innovation in breast cancer diagnostics and treatment planning.
Report of the Medical Image De-Identification Task Group -- Best Practices and Recommendations
Apr 01, 2023Abstract:This report addresses the technical aspects of de-identification of medical images of human subjects and biospecimens, such that re-identification risk of ethical, moral, and legal concern is sufficiently reduced to allow unrestricted public sharing for any purpose, regardless of the jurisdiction of the source and distribution sites. All medical images, regardless of the mode of acquisition, are considered, though the primary emphasis is on those with accompanying data elements, especially those encoded in formats in which the data elements are embedded, particularly Digital Imaging and Communications in Medicine (DICOM). These images include image-like objects such as Segmentations, Parametric Maps, and Radiotherapy (RT) Dose objects. The scope also includes related non-image objects, such as RT Structure Sets, Plans and Dose Volume Histograms, Structured Reports, and Presentation States. Only de-identification of publicly released data is considered, and alternative approaches to privacy preservation, such as federated learning for artificial intelligence (AI) model development, are out of scope, as are issues of privacy leakage from AI model sharing. Only technical issues of public sharing are addressed.
medigan: A Python Library of Pretrained Generative Models for Enriched Data Access in Medical Imaging
Sep 28, 2022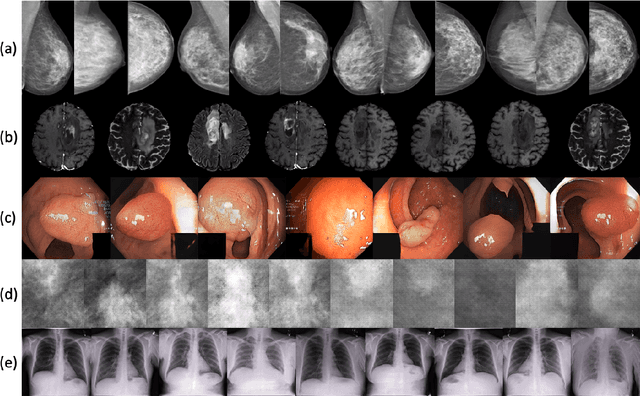
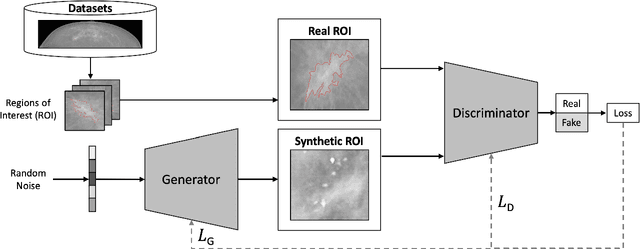
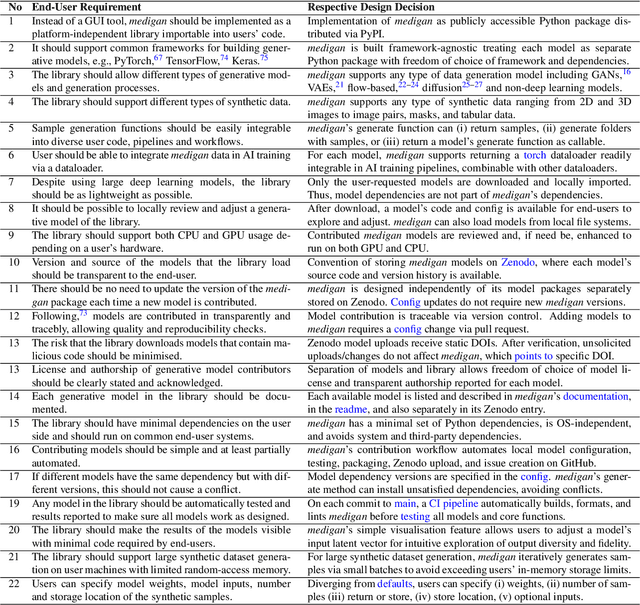
Abstract:Synthetic data generated by generative models can enhance the performance and capabilities of data-hungry deep learning models in medical imaging. However, there is (1) limited availability of (synthetic) datasets and (2) generative models are complex to train, which hinders their adoption in research and clinical applications. To reduce this entry barrier, we propose medigan, a one-stop shop for pretrained generative models implemented as an open-source framework-agnostic Python library. medigan allows researchers and developers to create, increase, and domain-adapt their training data in just a few lines of code. Guided by design decisions based on gathered end-user requirements, we implement medigan based on modular components for generative model (i) execution, (ii) visualisation, (iii) search & ranking, and (iv) contribution. The library's scalability and design is demonstrated by its growing number of integrated and readily-usable pretrained generative models consisting of 21 models utilising 9 different Generative Adversarial Network architectures trained on 11 datasets from 4 domains, namely, mammography, endoscopy, x-ray, and MRI. Furthermore, 3 applications of medigan are analysed in this work, which include (a) enabling community-wide sharing of restricted data, (b) investigating generative model evaluation metrics, and (c) improving clinical downstream tasks. In (b), extending on common medical image synthesis assessment and reporting standards, we show Fr\'echet Inception Distance variability based on image normalisation and radiology-specific feature extraction.
Lung cancer screening with low-dose CT scans using a deep learning approach
Jun 01, 2019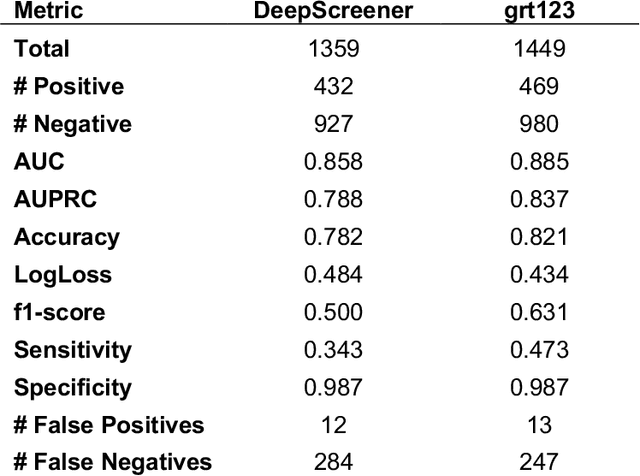
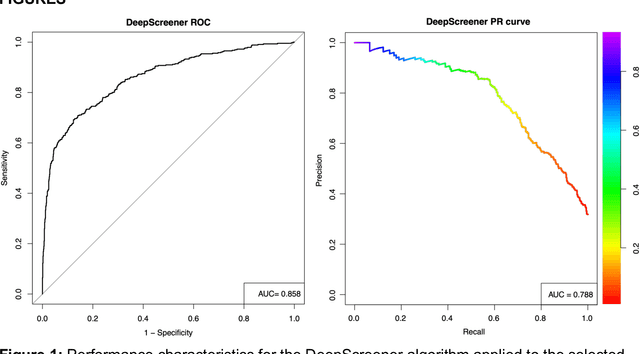
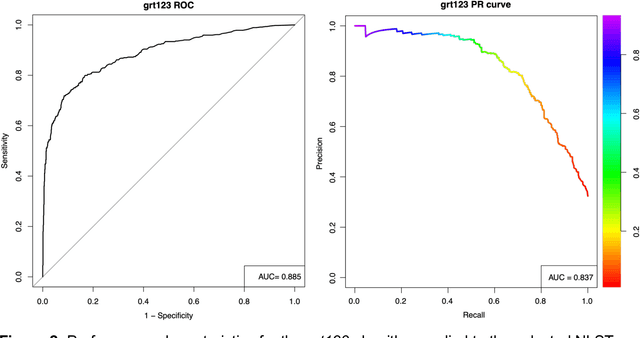
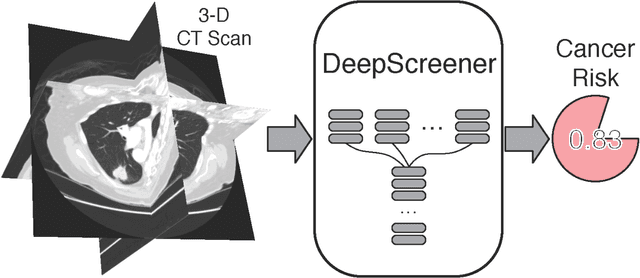
Abstract:Lung cancer is the leading cause of cancer deaths. Early detection through low-dose computed tomography (CT) screening has been shown to significantly reduce mortality but suffers from a high false positive rate that leads to unnecessary diagnostic procedures. Quantitative image analysis coupled to deep learning techniques has the potential to reduce this false positive rate. We conducted a computational analysis of 1449 low-dose CT studies drawn from the National Lung Screening Trial (NLST) cohort. We applied to this cohort our newly developed algorithm, DeepScreener, which is based on a novel deep learning approach. The algorithm, after the training process using about 3000 CT studies, does not require lung nodule annotations to conduct cancer prediction. The algorithm uses consecutive slices and multi-task features to determine whether a nodule is likely to be cancer, and a spatial pyramid to detect nodules at different scales. We find that the algorithm can predict a patient's cancer status from a volumetric lung CT image with high accuracy (78.2%, with area under the Receiver Operating Characteristic curve (AUC) of 0.858). Our preliminary framework ranked 16th of 1972 teams (top 1%) in the Data Science Bowl 2017 (DSB2017) competition, based on the challenge datasets. We report here the application of DeepScreener on an independent NLST test set. This study indicates that the deep learning approach has the potential to significantly reduce the false positive rate in lung cancer screening with low-dose CT scans.
Highly accurate model for prediction of lung nodule malignancy with CT scans
Feb 06, 2018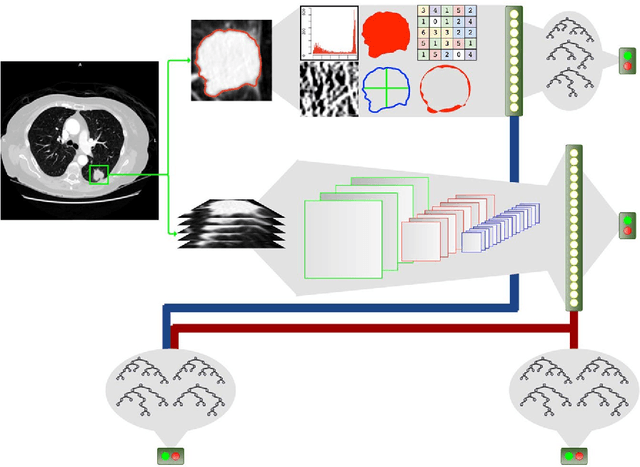
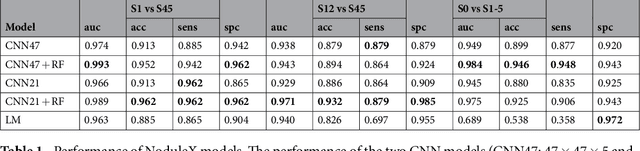
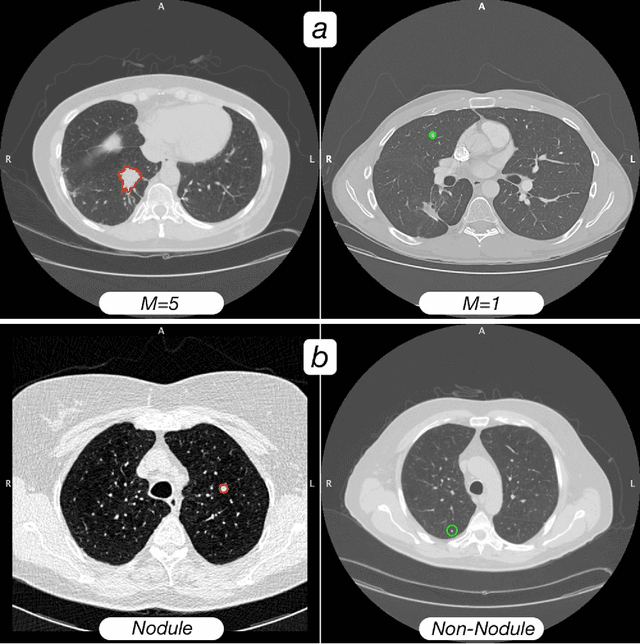
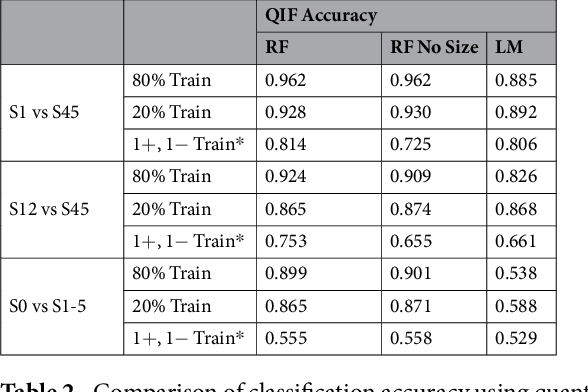
Abstract:Computed tomography (CT) examinations are commonly used to predict lung nodule malignancy in patients, which are shown to improve noninvasive early diagnosis of lung cancer. It remains challenging for computational approaches to achieve performance comparable to experienced radiologists. Here we present NoduleX, a systematic approach to predict lung nodule malignancy from CT data, based on deep learning convolutional neural networks (CNN). For training and validation, we analyze >1000 lung nodules in images from the LIDC/IDRI cohort. All nodules were identified and classified by four experienced thoracic radiologists who participated in the LIDC project. NoduleX achieves high accuracy for nodule malignancy classification, with an AUC of ~0.99. This is commensurate with the analysis of the dataset by experienced radiologists. Our approach, NoduleX, provides an effective framework for highly accurate nodule malignancy prediction with the model trained on a large patient population. Our results are replicable with software available at http://bioinformatics.astate.edu/NoduleX.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge