Jinglei Lv
Age Sensitive Hippocampal Functional Connectivity: New Insights from 3D CNNs and Saliency Mapping
Jul 02, 2025Abstract:Grey matter loss in the hippocampus is a hallmark of neurobiological aging, yet understanding the corresponding changes in its functional connectivity remains limited. Seed-based functional connectivity (FC) analysis enables voxel-wise mapping of the hippocampus's synchronous activity with cortical regions, offering a window into functional reorganization during aging. In this study, we develop an interpretable deep learning framework to predict brain age from hippocampal FC using a three-dimensional convolutional neural network (3D CNN) combined with LayerCAM saliency mapping. This approach maps key hippocampal-cortical connections, particularly with the precuneus, cuneus, posterior cingulate cortex, parahippocampal cortex, left superior parietal lobule, and right superior temporal sulcus, that are highly sensitive to age. Critically, disaggregating anterior and posterior hippocampal FC reveals distinct mapping aligned with their known functional specializations. These findings provide new insights into the functional mechanisms of hippocampal aging and demonstrate the power of explainable deep learning to uncover biologically meaningful patterns in neuroimaging data.
Voxel-Level Brain States Prediction Using Swin Transformer
Jun 13, 2025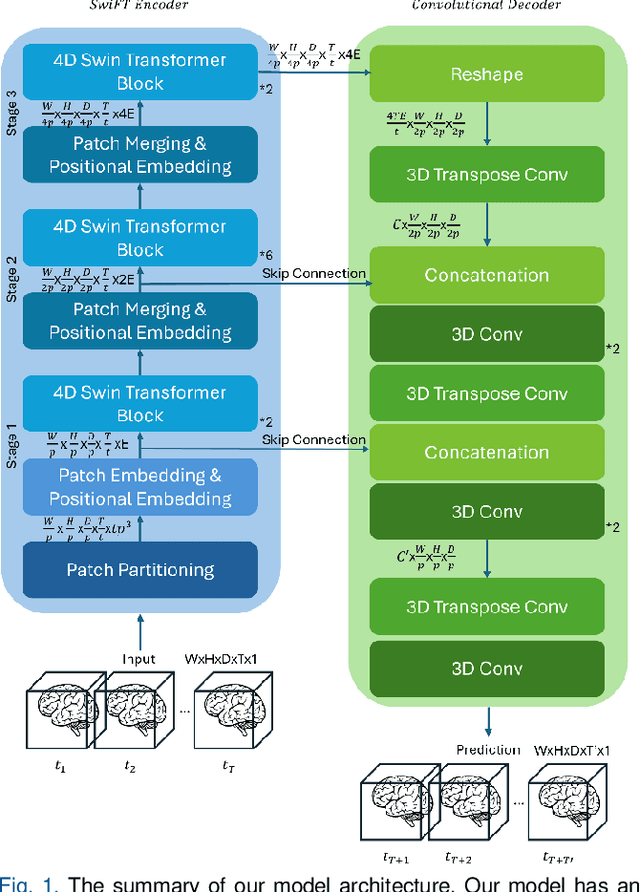

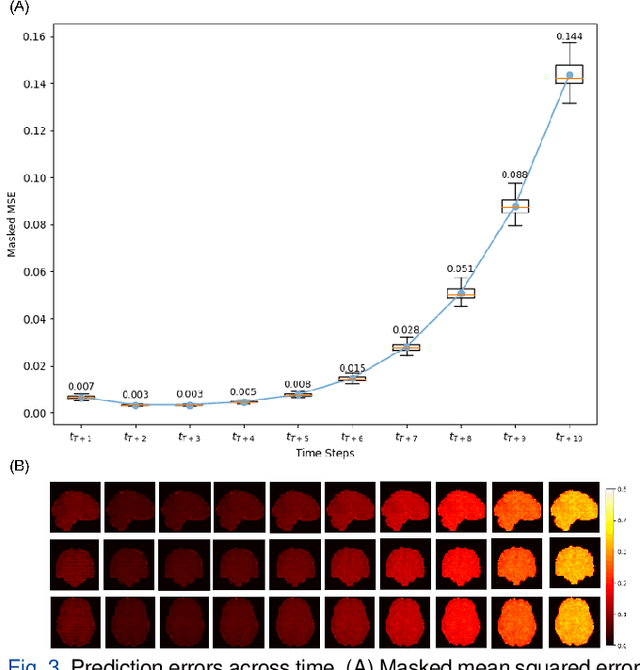

Abstract:Understanding brain dynamics is important for neuroscience and mental health. Functional magnetic resonance imaging (fMRI) enables the measurement of neural activities through blood-oxygen-level-dependent (BOLD) signals, which represent brain states. In this study, we aim to predict future human resting brain states with fMRI. Due to the 3D voxel-wise spatial organization and temporal dependencies of the fMRI data, we propose a novel architecture which employs a 4D Shifted Window (Swin) Transformer as encoder to efficiently learn spatio-temporal information and a convolutional decoder to enable brain state prediction at the same spatial and temporal resolution as the input fMRI data. We used 100 unrelated subjects from the Human Connectome Project (HCP) for model training and testing. Our novel model has shown high accuracy when predicting 7.2s resting-state brain activities based on the prior 23.04s fMRI time series. The predicted brain states highly resemble BOLD contrast and dynamics. This work shows promising evidence that the spatiotemporal organization of the human brain can be learned by a Swin Transformer model, at high resolution, which provides a potential for reducing the fMRI scan time and the development of brain-computer interfaces in the future.
Towards a general-purpose foundation model for fMRI analysis
Jun 11, 2025



Abstract:Functional Magnetic Resonance Imaging (fMRI) is essential for studying brain function and diagnosing neurological disorders, but current analysis methods face reproducibility and transferability issues due to complex pre-processing and task-specific models. We introduce NeuroSTORM (Neuroimaging Foundation Model with Spatial-Temporal Optimized Representation Modeling), a generalizable framework that directly learns from 4D fMRI volumes and enables efficient knowledge transfer across diverse applications. NeuroSTORM is pre-trained on 28.65 million fMRI frames (>9,000 hours) from over 50,000 subjects across multiple centers and ages 5 to 100. Using a Mamba backbone and a shifted scanning strategy, it efficiently processes full 4D volumes. We also propose a spatial-temporal optimized pre-training approach and task-specific prompt tuning to improve transferability. NeuroSTORM outperforms existing methods across five tasks: age/gender prediction, phenotype prediction, disease diagnosis, fMRI-to-image retrieval, and task-based fMRI classification. It demonstrates strong clinical utility on datasets from hospitals in the U.S., South Korea, and Australia, achieving top performance in disease diagnosis and cognitive phenotype prediction. NeuroSTORM provides a standardized, open-source foundation model to improve reproducibility and transferability in fMRI-based clinical research.
GCNs-Net: A Graph Convolutional Neural Network Approach for Decoding Time-resolved EEG Motor Imagery Signals
Jun 16, 2020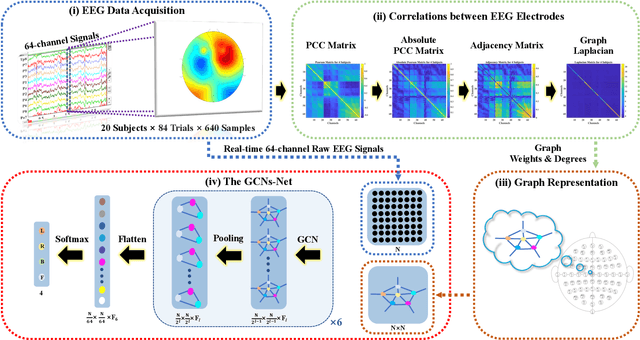
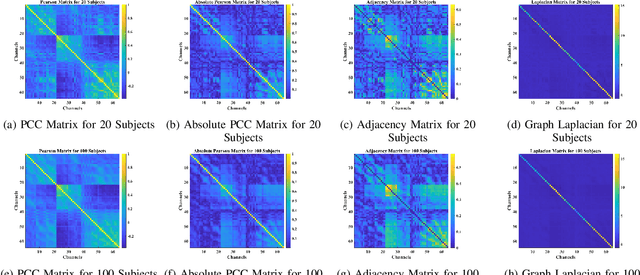
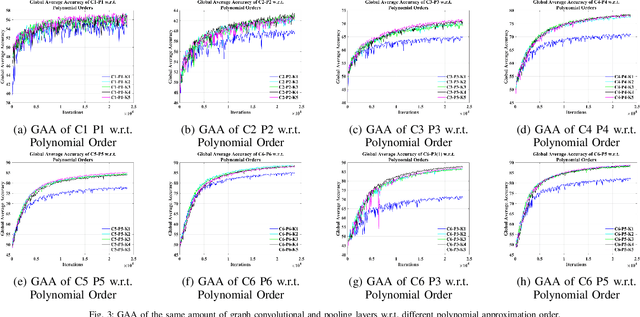
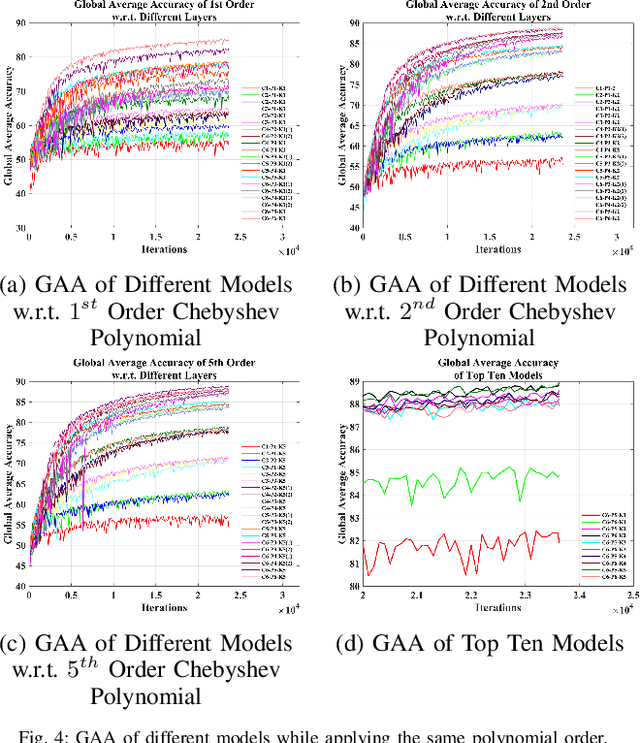
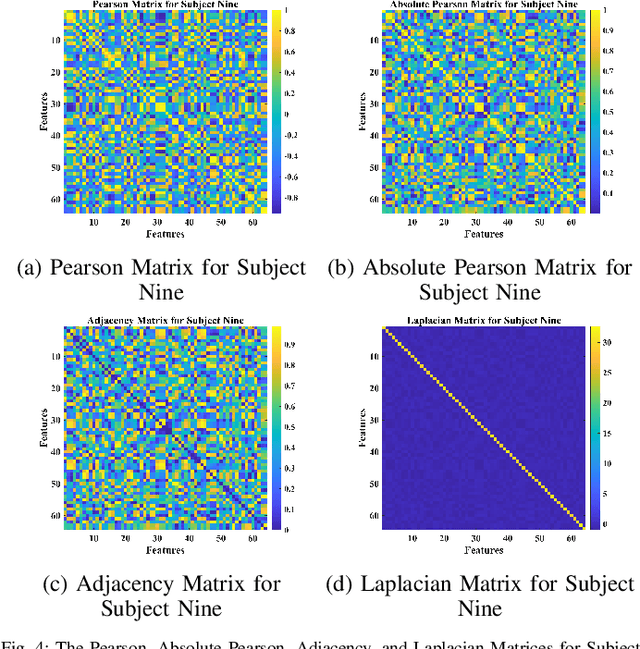
Abstract:Towards developing effective and efficient brain-computer interface (BCI) systems, precise decoding of brain activity measured by electroencephalogram (EEG), is highly demanded. Traditional works classify EEG signals without considering the topological relationship among electrodes. However, neuroscience research has increasingly emphasized network patterns of brain dynamics. Thus, the Euclidean structure of electrodes might not adequately reflect the interaction between signals. To fill the gap, a novel deep learning framework based on the graph convolutional neural networks (GCNs) was presented to enhance the decoding performance of raw EEG signals during different types of motor imagery (MI) tasks while cooperating with the functional topological relationship of electrodes. Based on the absolute Pearson's matrix of overall signals, the graph Laplacian of EEG electrodes was built up. The GCNs-Net constructed by graph convolutional layers learns the generalized features. The followed pooling layers reduce dimensionality, and the fully-connected softmax layer derives the final prediction. The introduced approach has been shown to converge for both personalized and group-wise predictions. It has achieved the highest averaged accuracy, 93.056% and 88.57% (PhysioNet Dataset), 96.24% and 80.89% (High Gamma Dataset), at the subject and group level, respectively, compared with existing studies, which suggests adaptability and robustness to individual variability. Moreover, the performance was stably reproducible among repetitive experiments for cross-validation. To conclude, the GCNs-Net filters EEG signals based on the functional topological relationship, which manages to decode relevant features for brain motor imagery.
Deep Feature Mining via Attention-based BiLSTM-GCN for Human Motor Imagery Recognition
May 02, 2020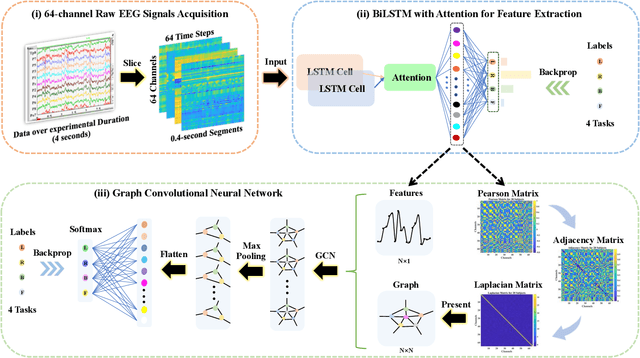
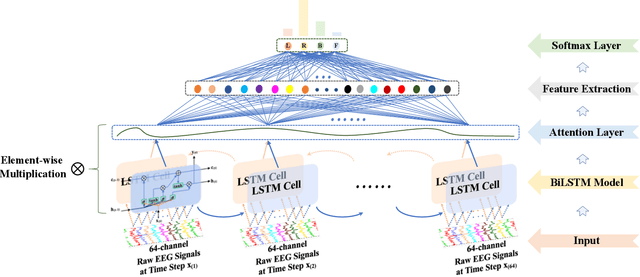
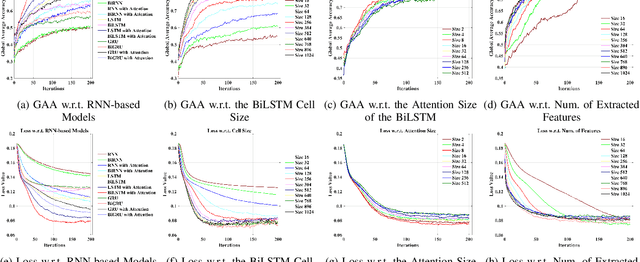
Abstract:Recognition accuracy and response time are both critically essential ahead of building practical electroencephalography (EEG) based brain-computer interface (BCI). Recent approaches, however, have either compromised in the classification accuracy or responding time. This paper presents a novel deep learning approach designed towards remarkably accurate and responsive motor imagery (MI) recognition based on scalp EEG. Bidirectional Long Short-term Memory (BiLSTM) with the Attention mechanism manages to derive relevant features from raw EEG signals. The connected graph convolutional neural network (GCN) promotes the decoding performance by cooperating with the topological structure of features, which are estimated from the overall data. The 0.4-second detection framework has shown effective and efficient prediction based on individual and group-wise training, with 98.81% and 94.64% accuracy, respectively, which outperformed all the state-of-the-art studies. The introduced deep feature mining approach can precisely recognize human motion intents from raw EEG signals, which paves the road to translate the EEG based MI recognition to practical BCI systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge