Jialin Ding
KramaBench: A Benchmark for AI Systems on Data-to-Insight Pipelines over Data Lakes
Jun 06, 2025Abstract:Constructing real-world data-to-insight pipelines often involves data extraction from data lakes, data integration across heterogeneous data sources, and diverse operations from data cleaning to analysis. The design and implementation of data science pipelines require domain knowledge, technical expertise, and even project-specific insights. AI systems have shown remarkable reasoning, coding, and understanding capabilities. However, it remains unclear to what extent these capabilities translate into successful design and execution of such complex pipelines. We introduce KRAMABENCH: a benchmark composed of 104 manually-curated real-world data science pipelines spanning 1700 data files from 24 data sources in 6 different domains. We show that these pipelines test the end-to-end capabilities of AI systems on data processing, requiring data discovery, wrangling and cleaning, efficient processing, statistical reasoning, and orchestrating data processing steps given a high-level task. Our evaluation tests 5 general models and 3 code generation models using our reference framework, DS-GURU, which instructs the AI model to decompose a question into a sequence of subtasks, reason through each step, and synthesize Python code that implements the proposed design. Our results on KRAMABENCH show that, although the models are sufficiently capable of solving well-specified data science code generation tasks, when extensive data processing and domain knowledge are required to construct real-world data science pipelines, existing out-of-box models fall short. Progress on KramaBench represents crucial steps towards developing autonomous data science agents for real-world applications. Our code, reference framework, and data are available at https://github.com/mitdbg/KramaBench.
TailorSQL: An NL2SQL System Tailored to Your Query Workload
May 29, 2025Abstract:NL2SQL (natural language to SQL) translates natural language questions into SQL queries, thereby making structured data accessible to non-technical users, serving as the foundation for intelligent data applications. State-of-the-art NL2SQL techniques typically perform translation by retrieving database-specific information, such as the database schema, and invoking a pre-trained large language model (LLM) using the question and retrieved information to generate the SQL query. However, existing NL2SQL techniques miss a key opportunity which is present in real-world settings: NL2SQL is typically applied on existing databases which have already served many SQL queries in the past. The past query workload implicitly contains information which is helpful for accurate NL2SQL translation and is not apparent from the database schema alone, such as common join paths and the semantics of obscurely-named tables and columns. We introduce TailorSQL, a NL2SQL system that takes advantage of information in the past query workload to improve both the accuracy and latency of translating natural language questions into SQL. By specializing to a given workload, TailorSQL achieves up to 2$\times$ improvement in execution accuracy on standardized benchmarks.
ODIN: A NL2SQL Recommender to Handle Schema Ambiguity
May 25, 2025Abstract:NL2SQL (natural language to SQL) systems translate natural language into SQL queries, allowing users with no technical background to interact with databases and create tools like reports or visualizations. While recent advancements in large language models (LLMs) have significantly improved NL2SQL accuracy, schema ambiguity remains a major challenge in enterprise environments with complex schemas, where multiple tables and columns with semantically similar names often co-exist. To address schema ambiguity, we introduce ODIN, a NL2SQL recommendation engine. Instead of producing a single SQL query given a natural language question, ODIN generates a set of potential SQL queries by accounting for different interpretations of ambiguous schema components. ODIN dynamically adjusts the number of suggestions based on the level of ambiguity, and ODIN learns from user feedback to personalize future SQL query recommendations. Our evaluation shows that ODIN improves the likelihood of generating the correct SQL query by 1.5-2$\times$ compared to baselines.
High-Order Associative Learning Based on Memristive Circuits for Efficient Learning
Oct 22, 2024



Abstract:Memristive associative learning has gained significant attention for its ability to mimic fundamental biological learning mechanisms while maintaining system simplicity. In this work, we introduce a high-order memristive associative learning framework with a biologically realistic structure. By utilizing memristors as synaptic modules and their state information to bridge different orders of associative learning, our design effectively establishes associations between multiple stimuli and replicates the transient nature of high-order associative learning. In Pavlov's classical conditioning experiments, our design achieves a 230% improvement in learning efficiency compared to previous works, with memristor power consumption in the synaptic modules remaining below 11 {\mu}W. In large-scale image recognition tasks, we utilize a 20*20 memristor array to represent images, enabling the system to recognize and label test images with semantic information at 100% accuracy. This scalability across different tasks highlights the framework's potential for a wide range of applications, offering enhanced learning efficiency for current memristor-based neuromorphic systems.
On the Pinsker bound of inner product kernel regression in large dimensions
Sep 02, 2024

Abstract:Building on recent studies of large-dimensional kernel regression, particularly those involving inner product kernels on the sphere $\mathbb{S}^{d}$, we investigate the Pinsker bound for inner product kernel regression in such settings. Specifically, we address the scenario where the sample size $n$ is given by $\alpha d^{\gamma}(1+o_{d}(1))$ for some $\alpha, \gamma>0$. We have determined the exact minimax risk for kernel regression in this setting, not only identifying the minimax rate but also the exact constant, known as the Pinsker constant, associated with the excess risk.
The Case for Learned Spatial Indexes
Aug 24, 2020
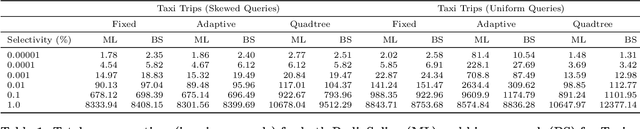
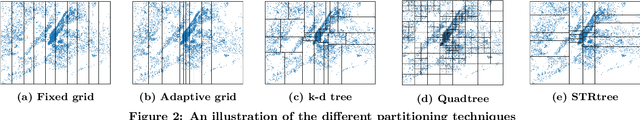
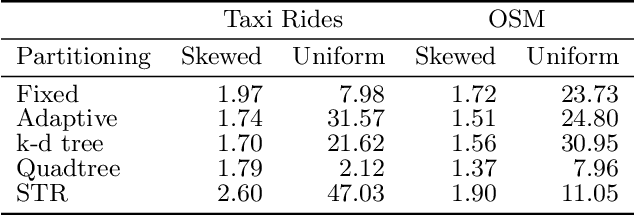
Abstract:Spatial data is ubiquitous. Massive amounts of data are generated every day from billions of GPS-enabled devices such as cell phones, cars, sensors, and various consumer-based applications such as Uber, Tinder, location-tagged posts in Facebook, Twitter, Instagram, etc. This exponential growth in spatial data has led the research community to focus on building systems and applications that can process spatial data efficiently. In the meantime, recent research has introduced learned index structures. In this work, we use techniques proposed from a state-of-the art learned multi-dimensional index structure (namely, Flood) and apply them to five classical multi-dimensional indexes to be able to answer spatial range queries. By tuning each partitioning technique for optimal performance, we show that (i) machine learned search within a partition is faster by 11.79\% to 39.51\% than binary search when using filtering on one dimension, (ii) the bottleneck for tree structures is index lookup, which could potentially be improved by linearizing the indexed partitions (iii) filtering on one dimension and refining using machine learned indexes is 1.23x to 1.83x times faster than closest competitor which filters on two dimensions, and (iv) learned indexes can have a significant impact on the performance of low selectivity queries while being less effective under higher selectivities.
Tsunami: A Learned Multi-dimensional Index for Correlated Data and Skewed Workloads
Jun 23, 2020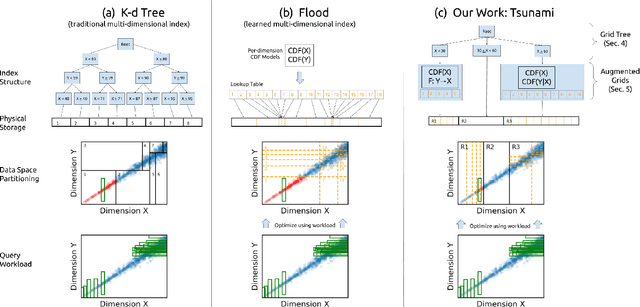
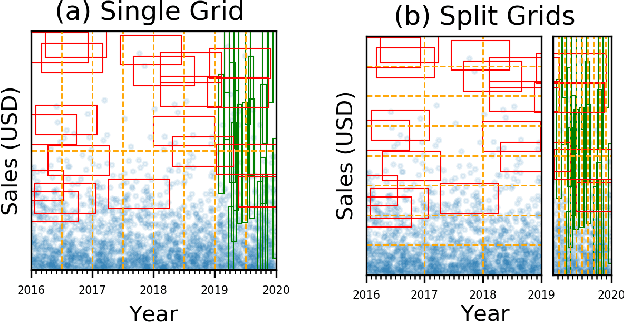

Abstract:Filtering data based on predicates is one of the most fundamental operations for any modern data warehouse. Techniques to accelerate the execution of filter expressions include clustered indexes, specialized sort orders (e.g., Z-order), multi-dimensional indexes, and, for high selectivity queries, secondary indexes. However, these schemes are hard to tune and their performance is inconsistent. Recent work on learned multi-dimensional indexes has introduced the idea of automatically optimizing an index for a particular dataset and workload. However, the performance of that work suffers in the presence of correlated data and skewed query workloads, both of which are common in real applications. In this paper, we introduce Tsunami, which addresses these limitations to achieve up to 6X faster query performance and up to 8X smaller index size than existing learned multi-dimensional indexes, in addition to up to 11X faster query performance and 170X smaller index size than optimally-tuned traditional indexes.
Learning Multi-dimensional Indexes
Dec 03, 2019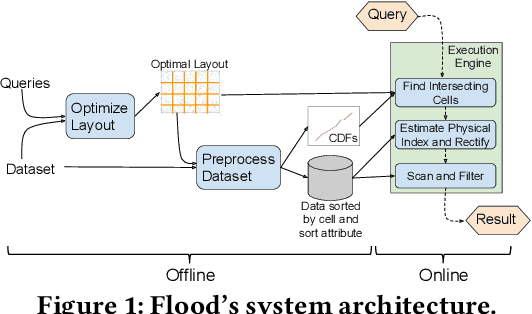
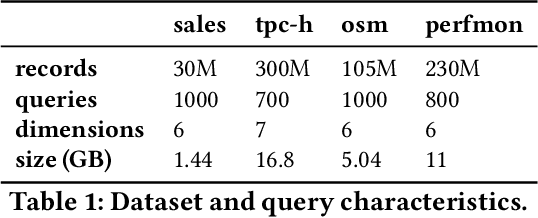
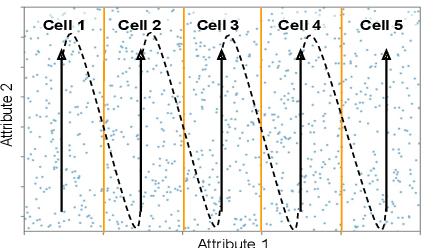
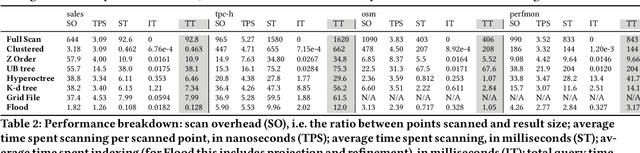
Abstract:Scanning and filtering over multi-dimensional tables are key operations in modern analytical database engines. To optimize the performance of these operations, databases often create clustered indexes over a single dimension or multi-dimensional indexes such as R-trees, or use complex sort orders (e.g., Z-ordering). However, these schemes are often hard to tune and their performance is inconsistent across different datasets and queries. In this paper, we introduce Flood, a multi-dimensional in-memory index that automatically adapts itself to a particular dataset and workload by jointly optimizing the index structure and data storage. Flood achieves up to three orders of magnitude faster performance for range scans with predicates than state-of-the-art multi-dimensional indexes or sort orders on real-world datasets and workloads. Our work serves as a building block towards an end-to-end learned database system.
LISA: Towards Learned DNA Sequence Search
Oct 10, 2019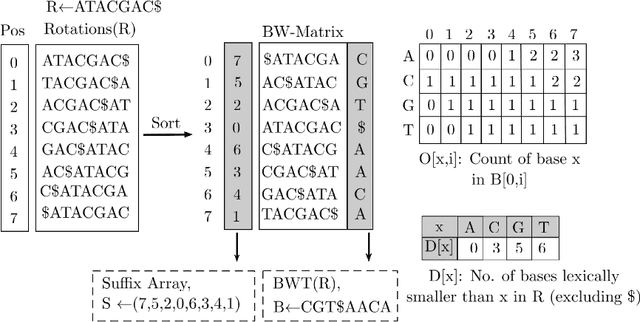

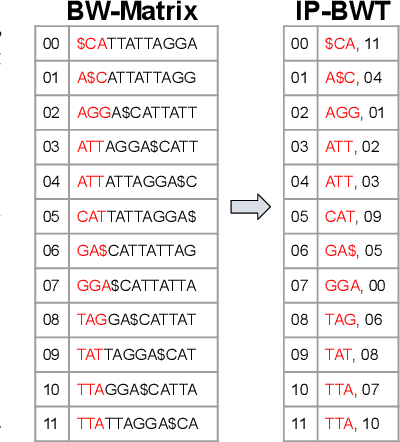
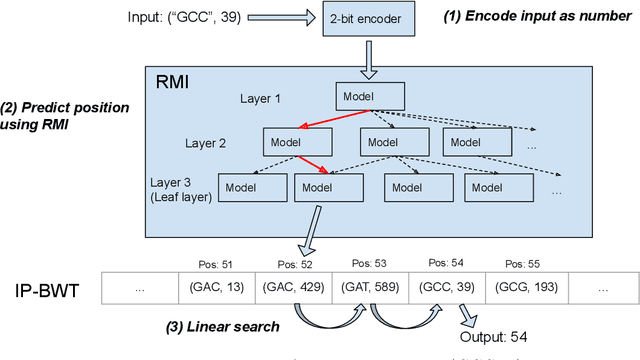
Abstract:Next-generation sequencing (NGS) technologies have enabled affordable sequencing of billions of short DNA fragments at high throughput, paving the way for population-scale genomics. Genomics data analytics at this scale requires overcoming performance bottlenecks, such as searching for short DNA sequences over long reference sequences. In this paper, we introduce LISA (Learned Indexes for Sequence Analysis), a novel learning-based approach to DNA sequence search. As a first proof of concept, we focus on accelerating one of the most essential flavors of the problem, called exact search. LISA builds on and extends FM-index, which is the state-of-the-art technique widely deployed in genomics tool-chains. Initial experiments with human genome datasets indicate that LISA achieves up to a factor of 4X performance speedup against its traditional counterpart.
ALEX: An Updatable Adaptive Learned Index
May 21, 2019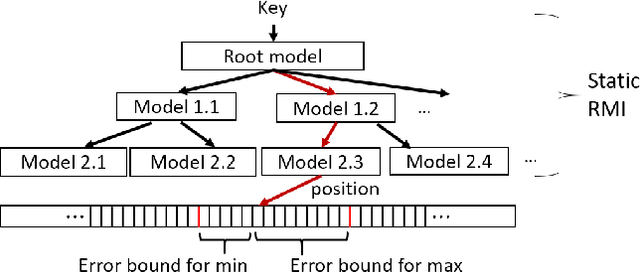
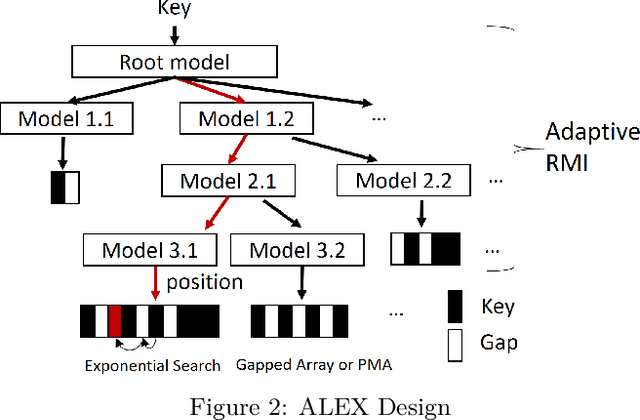
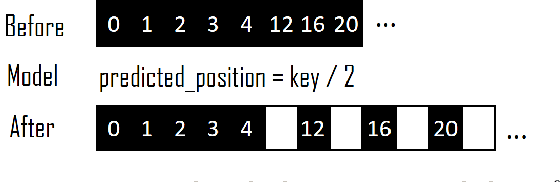
Abstract:Recent work on "learned indexes" has revolutionized the way we look at the decades-old field of DBMS indexing. The key idea is that indexes are "models" that predict the position of a key in a dataset. Indexes can, thus, be learned. The original work by Kraska et al. shows surprising results in terms of search performance and space requirements: A learned index beats a B+Tree by a factor of up to three in search time and by an order of magnitude in memory footprint, however it is limited to static, read-only workloads. This paper presents a new class of learned indexes called ALEX which addresses issues that arise when implementing dynamic, updatable learned indexes. Compared to the learned index from Kraska et al., ALEX has up to 3000X lower space requirements, but has up to 2.7X higher search performance on static workloads. Compared to a B+Tree, ALEX achieves up to 3.5X and 3.3X higher performance on static and some dynamic workloads, respectively, with up to 5 orders of magnitude smaller index size. Our detailed experiments show that ALEX presents a key step towards making learned indexes practical for a broader class of database workloads with dynamic updates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge