Darryl Ho
DejaVid: Encoder-Agnostic Learned Temporal Matching for Video Classification
Jun 14, 2025Abstract:In recent years, large transformer-based video encoder models have greatly advanced state-of-the-art performance on video classification tasks. However, these large models typically process videos by averaging embedding outputs from multiple clips over time to produce fixed-length representations. This approach fails to account for a variety of time-related features, such as variable video durations, chronological order of events, and temporal variance in feature significance. While methods for temporal modeling do exist, they often require significant architectural changes and expensive retraining, making them impractical for off-the-shelf, fine-tuned large encoders. To overcome these limitations, we propose DejaVid, an encoder-agnostic method that enhances model performance without the need for retraining or altering the architecture. Our framework converts a video into a variable-length temporal sequence of embeddings, which we call a multivariate time series (MTS). An MTS naturally preserves temporal order and accommodates variable video durations. We then learn per-timestep, per-feature weights over the encoded MTS frames, allowing us to account for variations in feature importance over time. We introduce a new neural network architecture inspired by traditional time series alignment algorithms for this learning task. Our evaluation demonstrates that DejaVid substantially improves the performance of a state-of-the-art large encoder, achieving leading Top-1 accuracy of 77.2% on Something-Something V2, 89.1% on Kinetics-400, and 88.6% on HMDB51, while adding fewer than 1.8% additional learnable parameters and requiring less than 3 hours of training time. Our code is available at https://github.com/darrylho/DejaVid.
LISA: Towards Learned DNA Sequence Search
Oct 10, 2019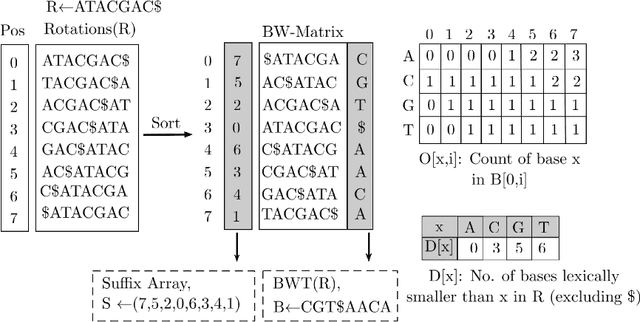

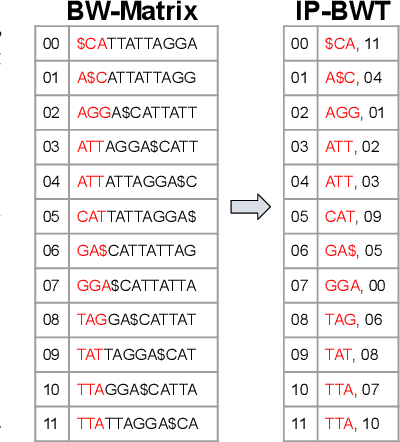
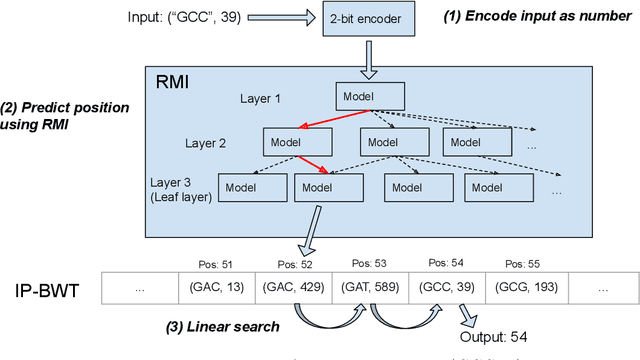
Abstract:Next-generation sequencing (NGS) technologies have enabled affordable sequencing of billions of short DNA fragments at high throughput, paving the way for population-scale genomics. Genomics data analytics at this scale requires overcoming performance bottlenecks, such as searching for short DNA sequences over long reference sequences. In this paper, we introduce LISA (Learned Indexes for Sequence Analysis), a novel learning-based approach to DNA sequence search. As a first proof of concept, we focus on accelerating one of the most essential flavors of the problem, called exact search. LISA builds on and extends FM-index, which is the state-of-the-art technique widely deployed in genomics tool-chains. Initial experiments with human genome datasets indicate that LISA achieves up to a factor of 4X performance speedup against its traditional counterpart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge