Huimin Huang
X-ray Insights Unleashed: Pioneering the Enhancement of Multi-Label Long-Tail Data
Dec 24, 2025Abstract:Long-tailed pulmonary anomalies in chest radiography present formidable diagnostic challenges. Despite the recent strides in diffusion-based methods for enhancing the representation of tailed lesions, the paucity of rare lesion exemplars curtails the generative capabilities of these approaches, thereby leaving the diagnostic precision less than optimal. In this paper, we propose a novel data synthesis pipeline designed to augment tail lesions utilizing a copious supply of conventional normal X-rays. Specifically, a sufficient quantity of normal samples is amassed to train a diffusion model capable of generating normal X-ray images. This pre-trained diffusion model is subsequently utilized to inpaint the head lesions present in the diseased X-rays, thereby preserving the tail classes as augmented training data. Additionally, we propose the integration of a Large Language Model Knowledge Guidance (LKG) module alongside a Progressive Incremental Learning (PIL) strategy to stabilize the inpainting fine-tuning process. Comprehensive evaluations conducted on the public lung datasets MIMIC and CheXpert demonstrate that the proposed method sets a new benchmark in performance.
BrainSegDMlF: A Dynamic Fusion-enhanced SAM for Brain Lesion Segmentation
May 09, 2025Abstract:The segmentation of substantial brain lesions is a significant and challenging task in the field of medical image segmentation. Substantial brain lesions in brain imaging exhibit high heterogeneity, with indistinct boundaries between lesion regions and normal brain tissue. Small lesions in single slices are difficult to identify, making the accurate and reproducible segmentation of abnormal regions, as well as their feature description, highly complex. Existing methods have the following limitations: 1) They rely solely on single-modal information for learning, neglecting the multi-modal information commonly used in diagnosis. This hampers the ability to comprehensively acquire brain lesion information from multiple perspectives and prevents the effective integration and utilization of multi-modal data inputs, thereby limiting a holistic understanding of lesions. 2) They are constrained by the amount of data available, leading to low sensitivity to small lesions and difficulty in detecting subtle pathological changes. 3) Current SAM-based models rely on external prompts, which cannot achieve automatic segmentation and, to some extent, affect diagnostic efficiency.To address these issues, we have developed a large-scale fully automated segmentation model specifically designed for brain lesion segmentation, named BrainSegDMLF. This model has the following features: 1) Dynamic Modal Interactive Fusion (DMIF) module that processes and integrates multi-modal data during the encoding process, providing the SAM encoder with more comprehensive modal information. 2) Layer-by-Layer Upsampling Decoder, enabling the model to extract rich low-level and high-level features even with limited data, thereby detecting the presence of small lesions. 3) Automatic segmentation masks, allowing the model to generate lesion masks automatically without requiring manual prompts.
ScaleFormer: Revisiting the Transformer-based Backbones from a Scale-wise Perspective for Medical Image Segmentation
Jul 29, 2022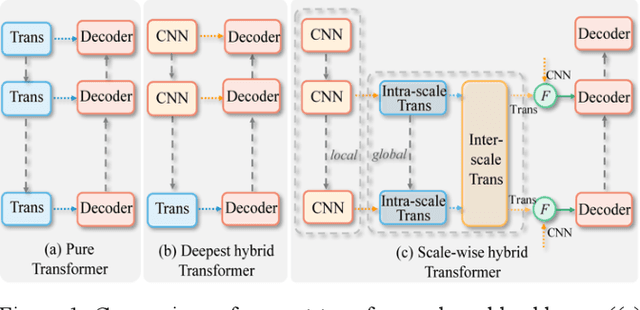
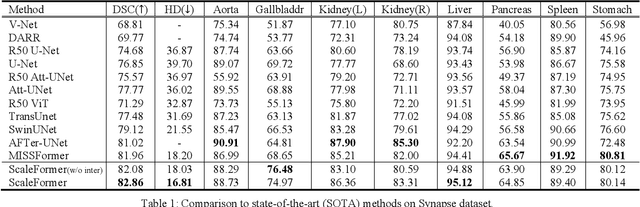
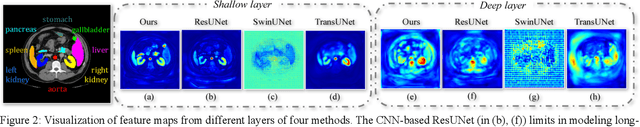
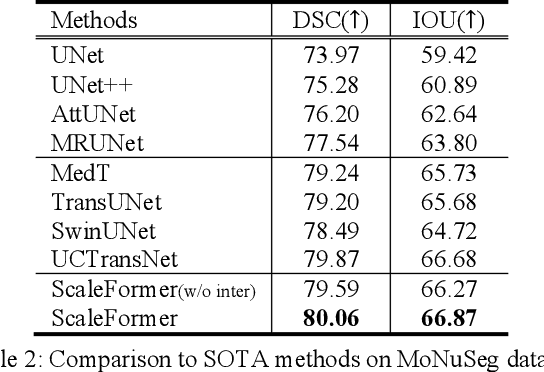
Abstract:Recently, a variety of vision transformers have been developed as their capability of modeling long-range dependency. In current transformer-based backbones for medical image segmentation, convolutional layers were replaced with pure transformers, or transformers were added to the deepest encoder to learn global context. However, there are mainly two challenges in a scale-wise perspective: (1) intra-scale problem: the existing methods lacked in extracting local-global cues in each scale, which may impact the signal propagation of small objects; (2) inter-scale problem: the existing methods failed to explore distinctive information from multiple scales, which may hinder the representation learning from objects with widely variable size, shape and location. To address these limitations, we propose a novel backbone, namely ScaleFormer, with two appealing designs: (1) A scale-wise intra-scale transformer is designed to couple the CNN-based local features with the transformer-based global cues in each scale, where the row-wise and column-wise global dependencies can be extracted by a lightweight Dual-Axis MSA. (2) A simple and effective spatial-aware inter-scale transformer is designed to interact among consensual regions in multiple scales, which can highlight the cross-scale dependency and resolve the complex scale variations. Experimental results on different benchmarks demonstrate that our Scale-Former outperforms the current state-of-the-art methods. The code is publicly available at: https://github.com/ZJUGiveLab/ScaleFormer.
Multi-phase Liver Tumor Segmentation with Spatial Aggregation and Uncertain Region Inpainting
Aug 05, 2021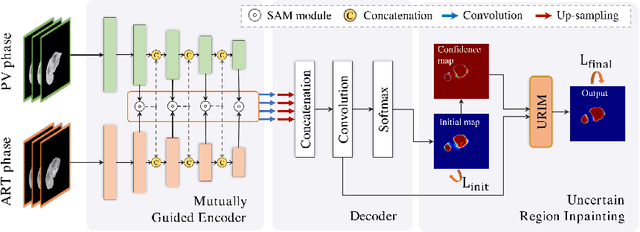
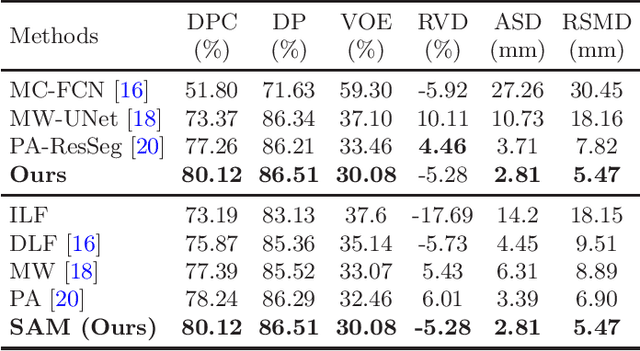
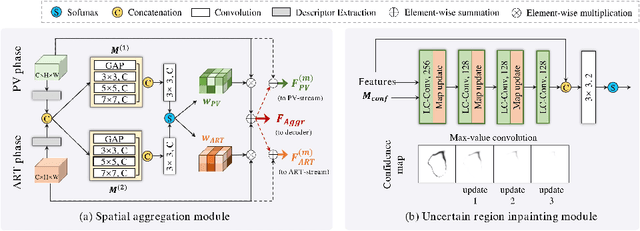
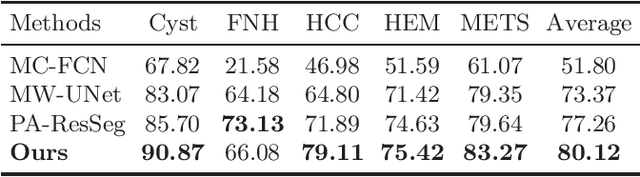
Abstract:Multi-phase computed tomography (CT) images provide crucial complementary information for accurate liver tumor segmentation (LiTS). State-of-the-art multi-phase LiTS methods usually fused cross-phase features through phase-weighted summation or channel-attention based concatenation. However, these methods ignored the spatial (pixel-wise) relationships between different phases, hence leading to insufficient feature integration. In addition, the performance of existing methods remains subject to the uncertainty in segmentation, which is particularly acute in tumor boundary regions. In this work, we propose a novel LiTS method to adequately aggregate multi-phase information and refine uncertain region segmentation. To this end, we introduce a spatial aggregation module (SAM), which encourages per-pixel interactions between different phases, to make full use of cross-phase information. Moreover, we devise an uncertain region inpainting module (URIM) to refine uncertain pixels using neighboring discriminative features. Experiments on an in-house multi-phase CT dataset of focal liver lesions (MPCT-FLLs) demonstrate that our method achieves promising liver tumor segmentation and outperforms state-of-the-arts.
Graph-based Pyramid Global Context Reasoning with a Saliency-aware Projection for COVID-19 Lung Infections Segmentation
Mar 07, 2021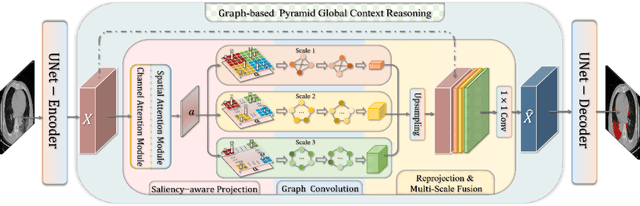
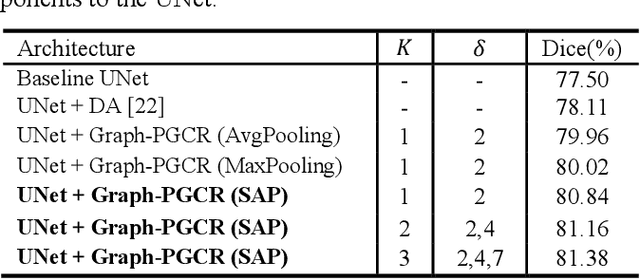
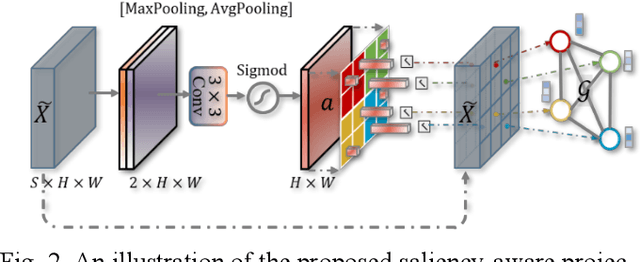
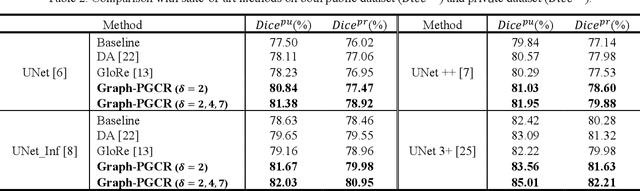
Abstract:Coronavirus Disease 2019 (COVID-19) has rapidly spread in 2020, emerging a mass of studies for lung infection segmentation from CT images. Though many methods have been proposed for this issue, it is a challenging task because of infections of various size appearing in different lobe zones. To tackle these issues, we propose a Graph-based Pyramid Global Context Reasoning (Graph-PGCR) module, which is capable of modeling long-range dependencies among disjoint infections as well as adapt size variation. We first incorporate graph convolution to exploit long-term contextual information from multiple lobe zones. Different from previous average pooling or maximum object probability, we propose a saliency-aware projection mechanism to pick up infection-related pixels as a set of graph nodes. After graph reasoning, the relation-aware features are reversed back to the original coordinate space for the down-stream tasks. We further construct multiple graphs with different sampling rates to handle the size variation problem. To this end, distinct multi-scale long-range contextual patterns can be captured. Our Graph-PGCR module is plug-and-play, which can be integrated into any architecture to improve its performance. Experiments demonstrated that the proposed method consistently boost the performance of state-of-the-art backbone architectures on both of public and our private COVID-19 datasets.
UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation
Apr 19, 2020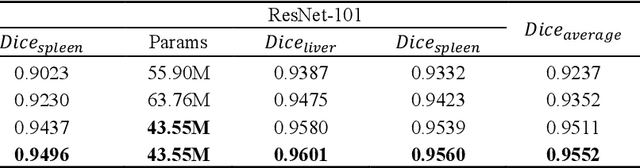
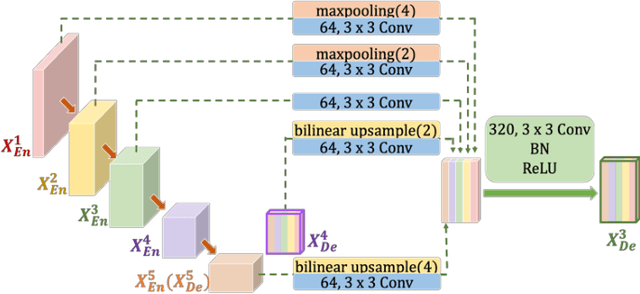
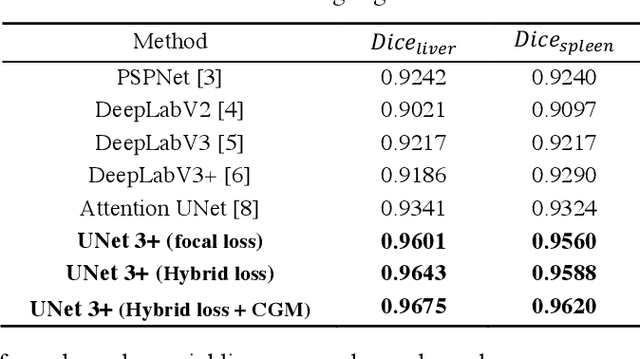
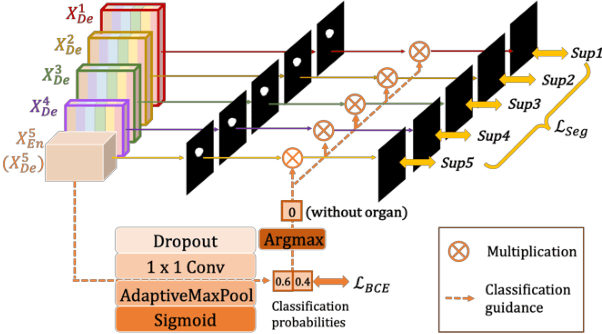
Abstract:Recently, a growing interest has been seen in deep learning-based semantic segmentation. UNet, which is one of deep learning networks with an encoder-decoder architecture, is widely used in medical image segmentation. Combining multi-scale features is one of important factors for accurate segmentation. UNet++ was developed as a modified Unet by designing an architecture with nested and dense skip connections. However, it does not explore sufficient information from full scales and there is still a large room for improvement. In this paper, we propose a novel UNet 3+, which takes advantage of full-scale skip connections and deep supervisions. The full-scale skip connections incorporate low-level details with high-level semantics from feature maps in different scales; while the deep supervision learns hierarchical representations from the full-scale aggregated feature maps. The proposed method is especially benefiting for organs that appear at varying scales. In addition to accuracy improvements, the proposed UNet 3+ can reduce the network parameters to improve the computation efficiency. We further propose a hybrid loss function and devise a classification-guided module to enhance the organ boundary and reduce the over-segmentation in a non-organ image, yielding more accurate segmentation results. The effectiveness of the proposed method is demonstrated on two datasets. The code is available at: github.com/ZJUGiveLab/UNet-Version
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge