Jia-Xuan Jiang
Vision-Language Semantic Aggregation Leveraging Foundation Model for Generalizable Medical Image Segmentation
Sep 10, 2025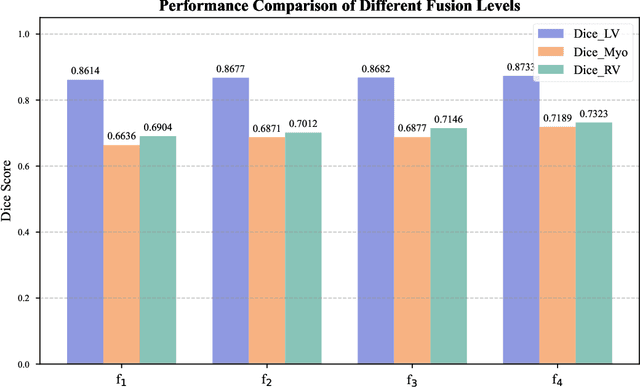
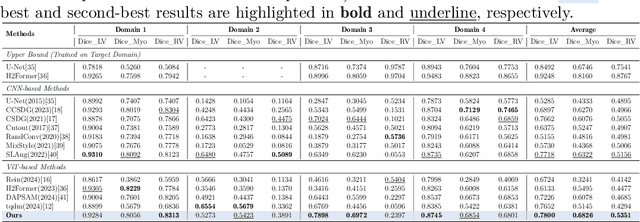
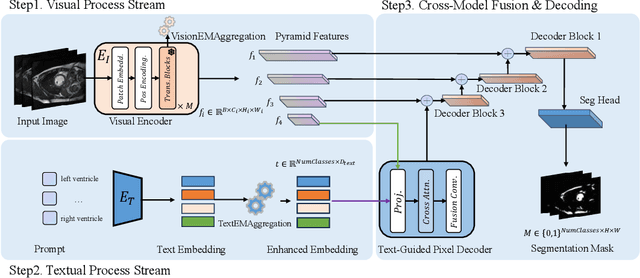
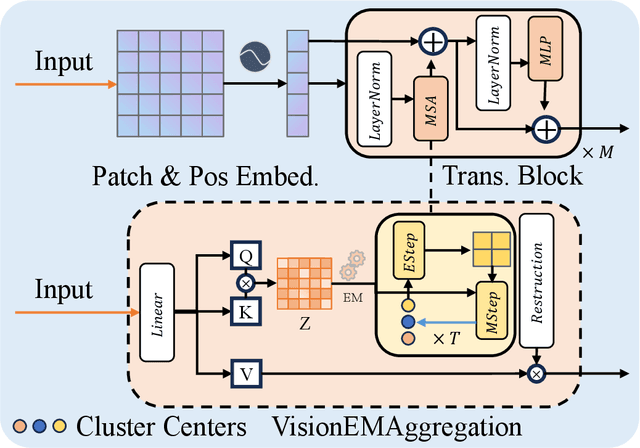
Abstract:Multimodal models have achieved remarkable success in natural image segmentation, yet they often underperform when applied to the medical domain. Through extensive study, we attribute this performance gap to the challenges of multimodal fusion, primarily the significant semantic gap between abstract textual prompts and fine-grained medical visual features, as well as the resulting feature dispersion. To address these issues, we revisit the problem from the perspective of semantic aggregation. Specifically, we propose an Expectation-Maximization (EM) Aggregation mechanism and a Text-Guided Pixel Decoder. The former mitigates feature dispersion by dynamically clustering features into compact semantic centers to enhance cross-modal correspondence. The latter is designed to bridge the semantic gap by leveraging domain-invariant textual knowledge to effectively guide deep visual representations. The synergy between these two mechanisms significantly improves the model's generalization ability. Extensive experiments on public cardiac and fundus datasets demonstrate that our method consistently outperforms existing SOTA approaches across multiple domain generalization benchmarks.
BrainSegDMlF: A Dynamic Fusion-enhanced SAM for Brain Lesion Segmentation
May 09, 2025Abstract:The segmentation of substantial brain lesions is a significant and challenging task in the field of medical image segmentation. Substantial brain lesions in brain imaging exhibit high heterogeneity, with indistinct boundaries between lesion regions and normal brain tissue. Small lesions in single slices are difficult to identify, making the accurate and reproducible segmentation of abnormal regions, as well as their feature description, highly complex. Existing methods have the following limitations: 1) They rely solely on single-modal information for learning, neglecting the multi-modal information commonly used in diagnosis. This hampers the ability to comprehensively acquire brain lesion information from multiple perspectives and prevents the effective integration and utilization of multi-modal data inputs, thereby limiting a holistic understanding of lesions. 2) They are constrained by the amount of data available, leading to low sensitivity to small lesions and difficulty in detecting subtle pathological changes. 3) Current SAM-based models rely on external prompts, which cannot achieve automatic segmentation and, to some extent, affect diagnostic efficiency.To address these issues, we have developed a large-scale fully automated segmentation model specifically designed for brain lesion segmentation, named BrainSegDMLF. This model has the following features: 1) Dynamic Modal Interactive Fusion (DMIF) module that processes and integrates multi-modal data during the encoding process, providing the SAM encoder with more comprehensive modal information. 2) Layer-by-Layer Upsampling Decoder, enabling the model to extract rich low-level and high-level features even with limited data, thereby detecting the presence of small lesions. 3) Automatic segmentation masks, allowing the model to generate lesion masks automatically without requiring manual prompts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge