Dimitris Samaras
SUNY
Learning Probabilistic Structural Representation for Biomedical Image Segmentation
Jun 03, 2022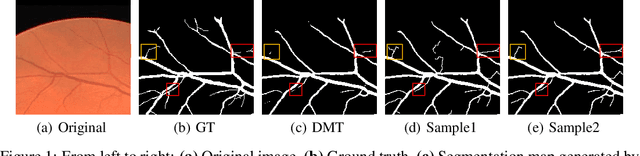
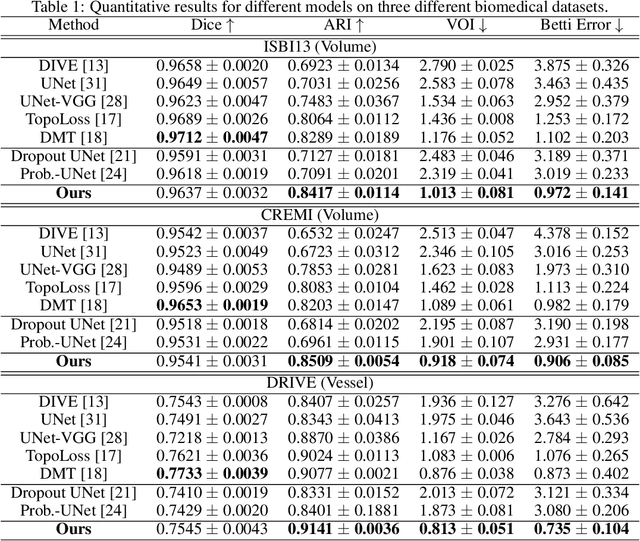
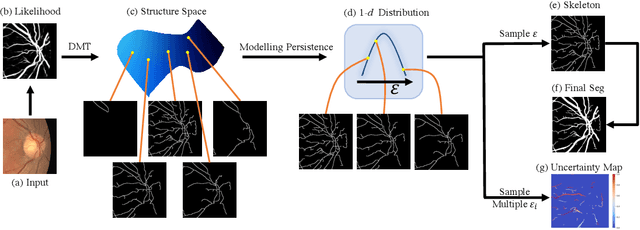
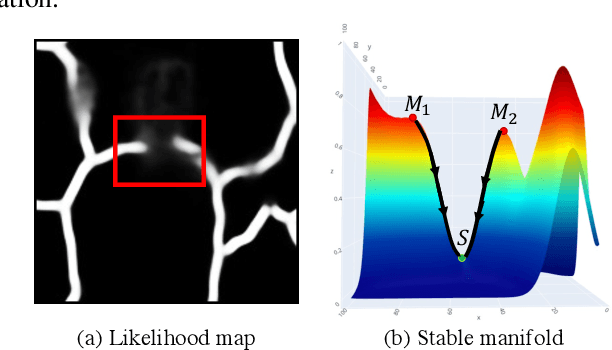
Abstract:Accurate segmentation of various fine-scale structures from biomedical images is a very important yet challenging problem. Existing methods use topological information as an additional training loss, but are ultimately learning a pixel-wise representation. In this paper, we propose the first deep learning method to learn a structural representation. We use discrete Morse theory and persistent homology to construct an one-parameter family of structures as the structural representation space. Furthermore, we learn a probabilistic model that can do inference tasks on such a structural representation space. We empirically demonstrate the strength of our method, i.e., generating true structures rather than pixel-maps with better topological integrity, and facilitating a human-in-the-loop annotation pipeline using the sampling of structures and structure-aware uncertainty.
A Novel Framework for Characterization of Tumor-Immune Spatial Relationships in Tumor Microenvironment
May 01, 2022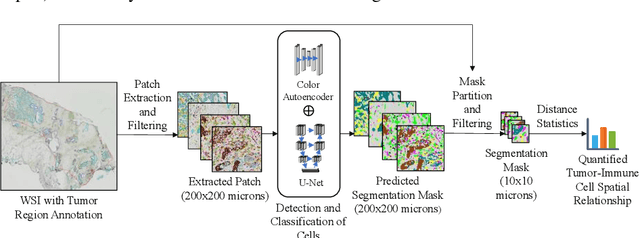

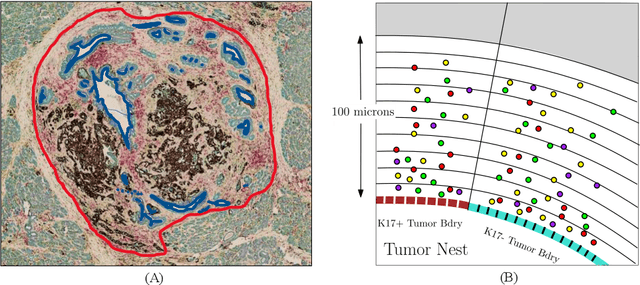
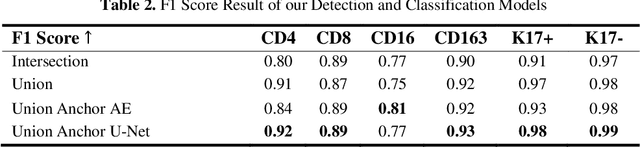
Abstract:Understanding the impact of tumor biology on the composition of nearby cells often requires characterizing the impact of biologically distinct tumor regions. Biomarkers have been developed to label biologically distinct tumor regions, but challenges arise because of differences in the spatial extent and distribution of differentially labeled regions. In this work, we present a framework for systematically investigating the impact of distinct tumor regions on cells near the tumor borders, accounting their cross spatial distributions. We apply the framework to multiplex immunohistochemistry (mIHC) studies of pancreatic cancer and show its efficacy in demonstrating how biologically different tumor regions impact the immune response in the tumor microenvironment. Furthermore, we show that the proposed framework can be extended to largescale whole slide image analysis.
Self Pre-training with Masked Autoencoders for Medical Image Analysis
Mar 10, 2022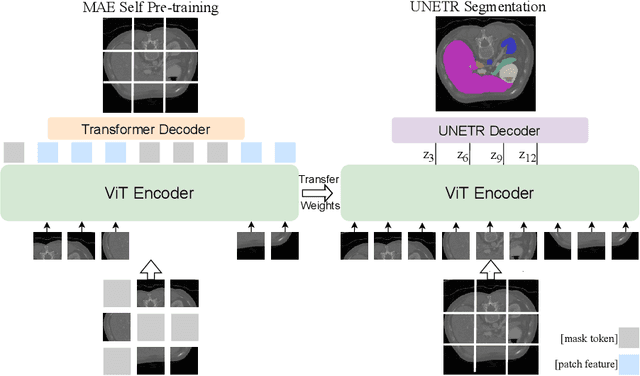
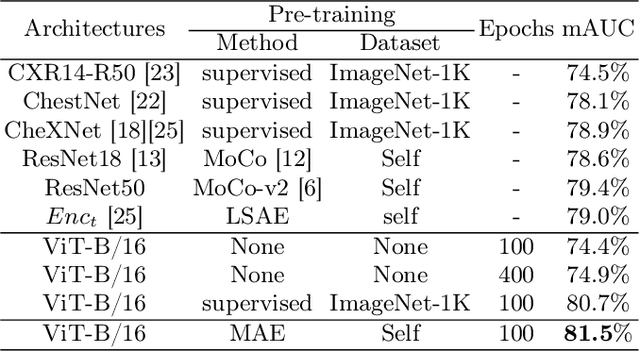
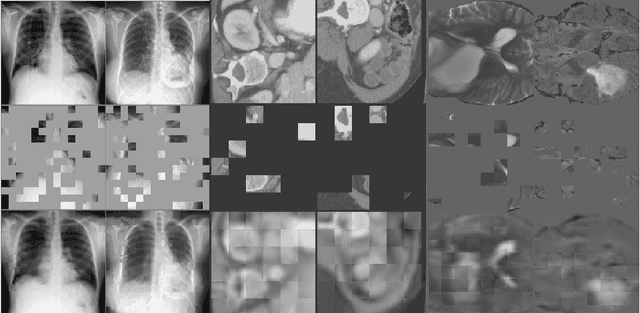
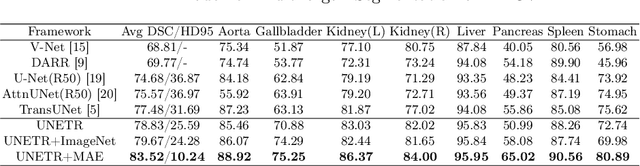
Abstract:Masked Autoencoder (MAE) has recently been shown to be effective in pre-training Vision Transformers (ViT) for natural image analysis. By performing the pretext task of reconstructing the original image from only partial observations, the encoder, which is a ViT, is encouraged to aggregate contextual information to infer content in masked image regions. We believe that this context aggregation ability is also essential to the medical image domain where each anatomical structure is functionally and mechanically connected to other structures and regions. However, there is no ImageNet-scale medical image dataset for pre-training. Thus, in this paper, we investigate a self pre-training paradigm with MAE for medical images, i.e., models are pre-trained on the same target dataset. To validate the MAE self pre-training, we consider three diverse medical image tasks including chest X-ray disease classification, CT abdomen multi-organ segmentation and MRI brain tumor segmentation. It turns out MAE self pre-training benefits all the tasks markedly. Specifically, the mAUC on lung disease classification is increased by 9.4%. The average DSC on brain tumor segmentation is improved from 77.4% to 78.9%. Most interestingly, on the small-scale multi-organ segmentation dataset (N=30), the average DSC improves from 78.8% to 83.5% and the HD95 is reduced by 60%, indicating its effectiveness in limited data scenarios. The segmentation and classification results reveal the promising potential of MAE self pre-training for medical image analysis.
Visual attention analysis of pathologists examining whole slide images of Prostate cancer
Feb 17, 2022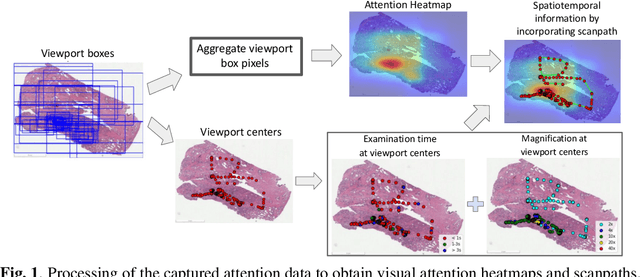
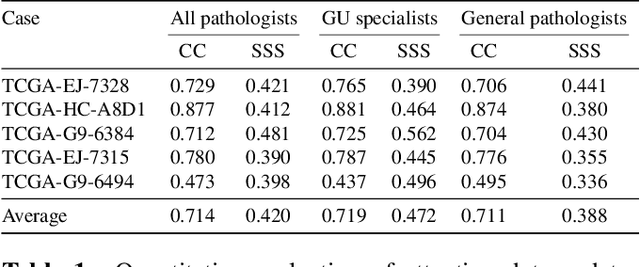
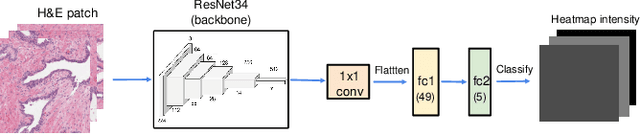
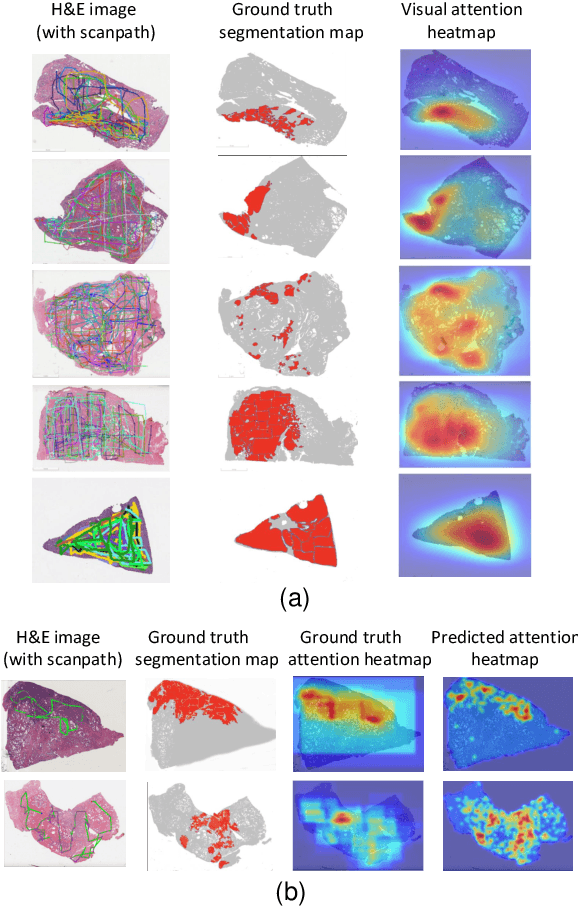
Abstract:We study the attention of pathologists as they examine whole-slide images (WSIs) of prostate cancer tissue using a digital microscope. To the best of our knowledge, our study is the first to report in detail how pathologists navigate WSIs of prostate cancer as they accumulate information for their diagnoses. We collected slide navigation data (i.e., viewport location, magnification level, and time) from 13 pathologists in 2 groups (5 genitourinary (GU) specialists and 8 general pathologists) and generated visual attention heatmaps and scanpaths. Each pathologist examined five WSIs from the TCGA PRAD dataset, which were selected by a GU pathology specialist. We examined and analyzed the distributions of visual attention for each group of pathologists after each WSI was examined. To quantify the relationship between a pathologist's attention and evidence for cancer in the WSI, we obtained tumor annotations from a genitourinary specialist. We used these annotations to compute the overlap between the distribution of visual attention and annotated tumor region to identify strong correlations. Motivated by this analysis, we trained a deep learning model to predict visual attention on unseen WSIs. We find that the attention heatmaps predicted by our model correlate quite well with the ground truth attention heatmap and tumor annotations on a test set of 17 WSIs by using various spatial and temporal evaluation metrics.
Lung Swapping Autoencoder: Learning a Disentangled Structure-texture Representation of Chest Radiographs
Jan 18, 2022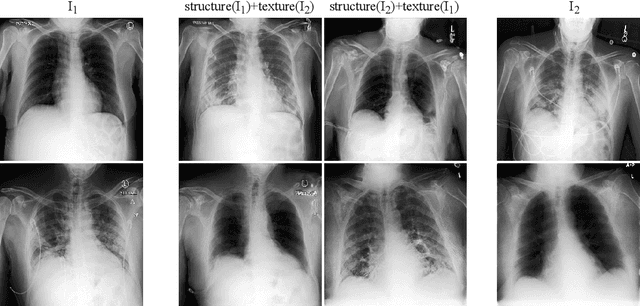

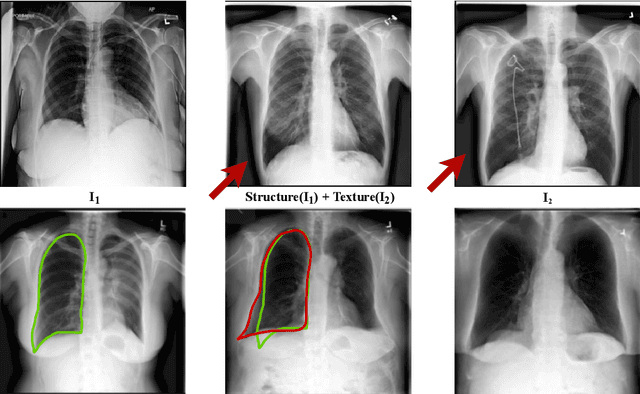
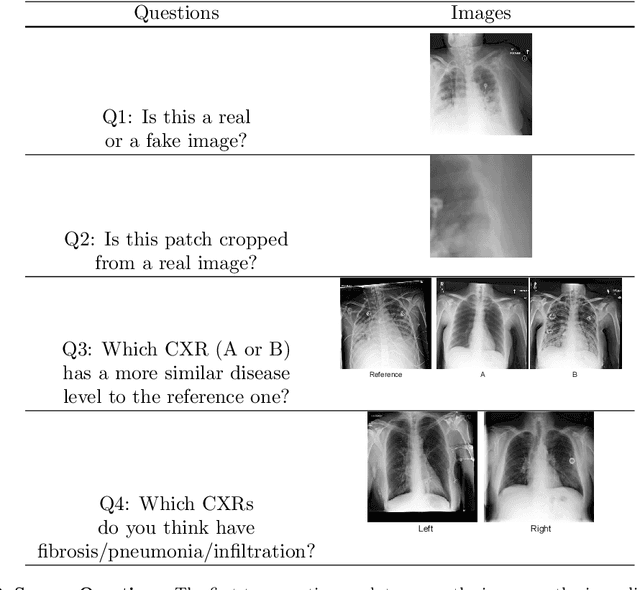
Abstract:Well-labeled datasets of chest radiographs (CXRs) are difficult to acquire due to the high cost of annotation. Thus, it is desirable to learn a robust and transferable representation in an unsupervised manner to benefit tasks that lack labeled data. Unlike natural images, medical images have their own domain prior; e.g., we observe that many pulmonary diseases, such as the COVID-19, manifest as changes in the lung tissue texture rather than the anatomical structure. Therefore, we hypothesize that studying only the texture without the influence of structure variations would be advantageous for downstream prognostic and predictive modeling tasks. In this paper, we propose a generative framework, the Lung Swapping Autoencoder (LSAE), that learns factorized representations of a CXR to disentangle the texture factor from the structure factor. Specifically, by adversarial training, the LSAE is optimized to generate a hybrid image that preserves the lung shape in one image but inherits the lung texture of another. To demonstrate the effectiveness of the disentangled texture representation, we evaluate the texture encoder $Enc^t$ in LSAE on ChestX-ray14 (N=112,120), and our own multi-institutional COVID-19 outcome prediction dataset, COVOC (N=340 (Subset-1) + 53 (Subset-2)). On both datasets, we reach or surpass the state-of-the-art by finetuning $Enc^t$ in LSAE that is 77% smaller than a baseline Inception v3. Additionally, in semi-and-self supervised settings with a similar model budget, $Enc^t$ in LSAE is also competitive with the state-of-the-art MoCo. By "re-mixing" the texture and shape factors, we generate meaningful hybrid images that can augment the training set. This data augmentation method can further improve COVOC prediction performance. The improvement is consistent even when we directly evaluate the Subset-1 trained model on Subset-2 without any fine-tuning.
CoDiM: Learning with Noisy Labels via Contrastive Semi-Supervised Learning
Nov 23, 2021
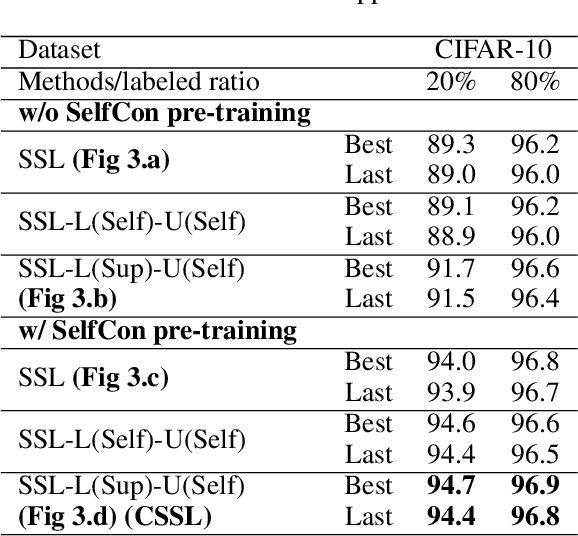
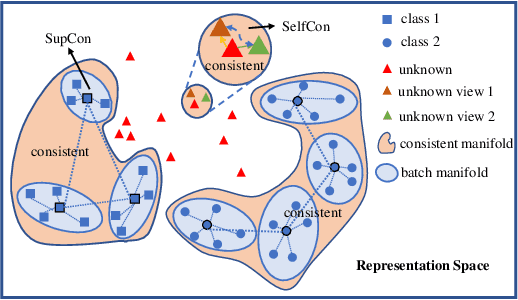
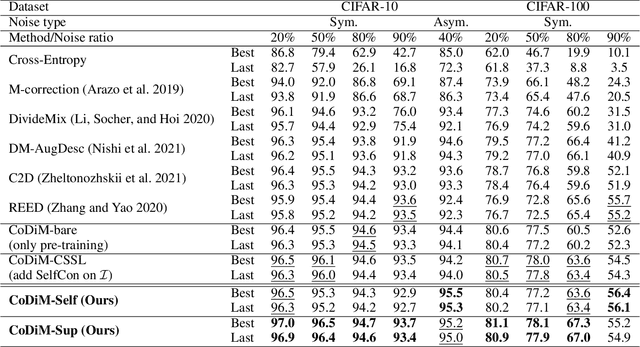
Abstract:Labels are costly and sometimes unreliable. Noisy label learning, semi-supervised learning, and contrastive learning are three different strategies for designing learning processes requiring less annotation cost. Semi-supervised learning and contrastive learning have been recently demonstrated to improve learning strategies that address datasets with noisy labels. Still, the inner connections between these fields as well as the potential to combine their strengths together have only started to emerge. In this paper, we explore further ways and advantages to fuse them. Specifically, we propose CSSL, a unified Contrastive Semi-Supervised Learning algorithm, and CoDiM (Contrastive DivideMix), a novel algorithm for learning with noisy labels. CSSL leverages the power of classical semi-supervised learning and contrastive learning technologies and is further adapted to CoDiM, which learns robustly from multiple types and levels of label noise. We show that CoDiM brings consistent improvements and achieves state-of-the-art results on multiple benchmarks.
Multi-Class Cell Detection Using Spatial Context Representation
Oct 10, 2021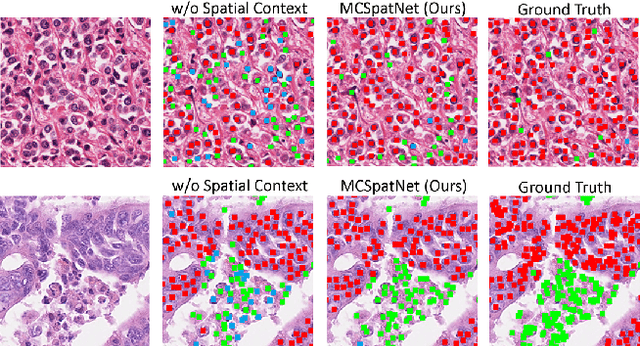

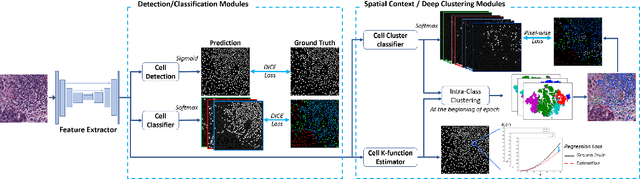
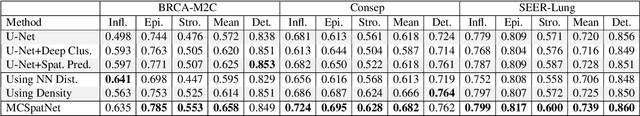
Abstract:In digital pathology, both detection and classification of cells are important for automatic diagnostic and prognostic tasks. Classifying cells into subtypes, such as tumor cells, lymphocytes or stromal cells is particularly challenging. Existing methods focus on morphological appearance of individual cells, whereas in practice pathologists often infer cell classes through their spatial context. In this paper, we propose a novel method for both detection and classification that explicitly incorporates spatial contextual information. We use the spatial statistical function to describe local density in both a multi-class and a multi-scale manner. Through representation learning and deep clustering techniques, we learn advanced cell representation with both appearance and spatial context. On various benchmarks, our method achieves better performance than state-of-the-arts, especially on the classification task.
SIDER: Single-Image Neural Optimization for Facial Geometric Detail Recovery
Aug 11, 2021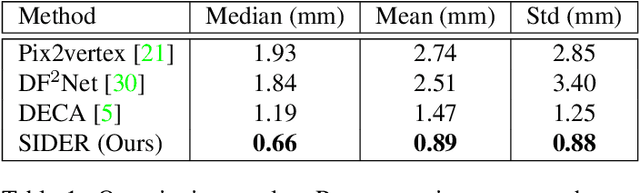
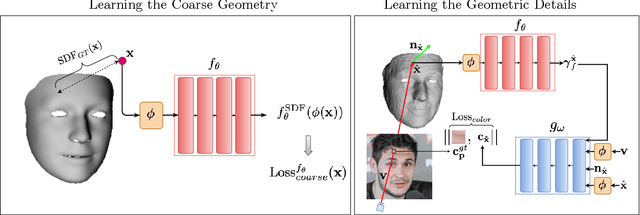


Abstract:We present SIDER(Single-Image neural optimization for facial geometric DEtail Recovery), a novel photometric optimization method that recovers detailed facial geometry from a single image in an unsupervised manner. Inspired by classical techniques of coarse-to-fine optimization and recent advances in implicit neural representations of 3D shape, SIDER combines a geometry prior based on statistical models and Signed Distance Functions (SDFs) to recover facial details from single images. First, it estimates a coarse geometry using a morphable model represented as an SDF. Next, it reconstructs facial geometry details by optimizing a photometric loss with respect to the ground truth image. In contrast to prior work, SIDER does not rely on any dataset priors and does not require additional supervision from multiple views, lighting changes or ground truth 3D shape. Extensive qualitative and quantitative evaluation demonstrates that our method achieves state-of-the-art on facial geometric detail recovery, using only a single in-the-wild image.
FLAME-in-NeRF : Neural control of Radiance Fields for Free View Face Animation
Aug 10, 2021



Abstract:This paper presents a neural rendering method for controllable portrait video synthesis. Recent advances in volumetric neural rendering, such as neural radiance fields (NeRF), has enabled the photorealistic novel view synthesis of static scenes with impressive results. However, modeling dynamic and controllable objects as part of a scene with such scene representations is still challenging. In this work, we design a system that enables both novel view synthesis for portrait video, including the human subject and the scene background, and explicit control of the facial expressions through a low-dimensional expression representation. We leverage the expression space of a 3D morphable face model (3DMM) to represent the distribution of human facial expressions, and use it to condition the NeRF volumetric function. Furthermore, we impose a spatial prior brought by 3DMM fitting to guide the network to learn disentangled control for scene appearance and facial actions. We demonstrate the effectiveness of our method on free view synthesis of portrait videos with expression controls. To train a scene, our method only requires a short video of a subject captured by a mobile device.
Temporal Feature Warping for Video Shadow Detection
Jul 29, 2021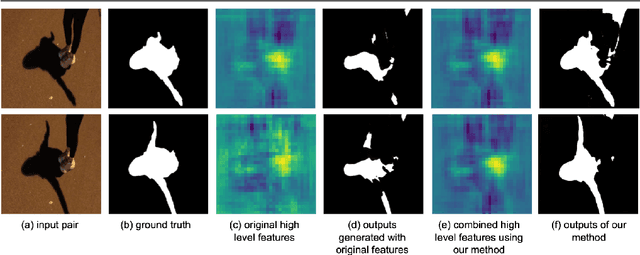
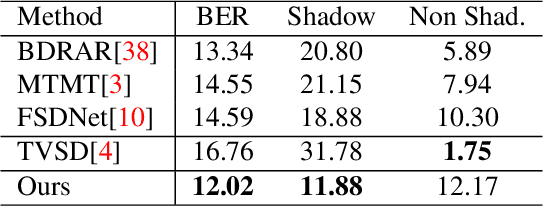
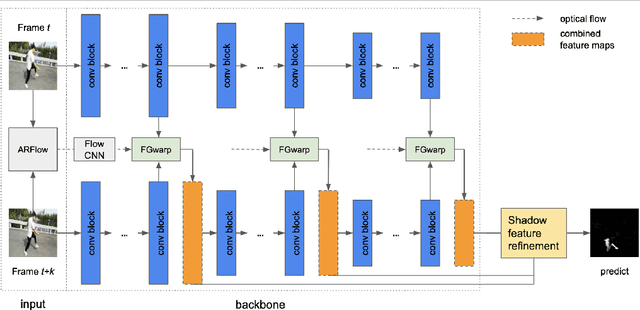
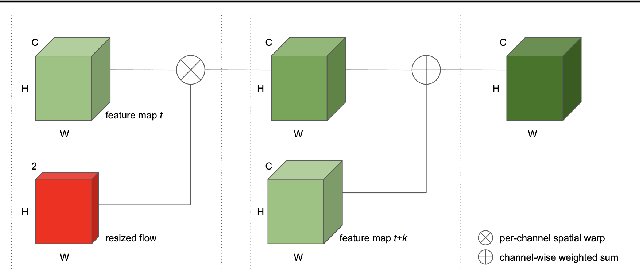
Abstract:While single image shadow detection has been improving rapidly in recent years, video shadow detection remains a challenging task due to data scarcity and the difficulty in modelling temporal consistency. The current video shadow detection method achieves this goal via co-attention, which mostly exploits information that is temporally coherent but is not robust in detecting moving shadows and small shadow regions. In this paper, we propose a simple but powerful method to better aggregate information temporally. We use an optical flow based warping module to align and then combine features between frames. We apply this warping module across multiple deep-network layers to retrieve information from neighboring frames including both local details and high-level semantic information. We train and test our framework on the ViSha dataset. Experimental results show that our model outperforms the state-of-the-art video shadow detection method by 28%, reducing BER from 16.7 to 12.0.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge