Buzhou Tang
From Long to Lean: Performance-aware and Adaptive Chain-of-Thought Compression via Multi-round Refinement
Sep 26, 2025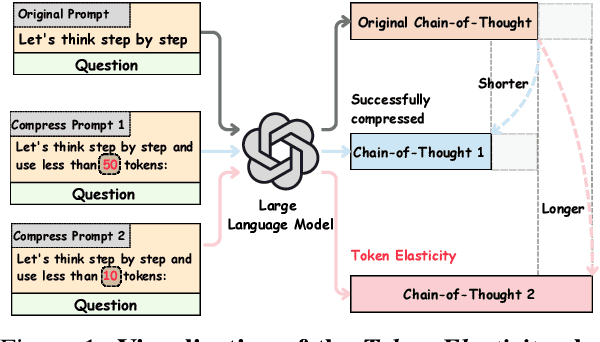
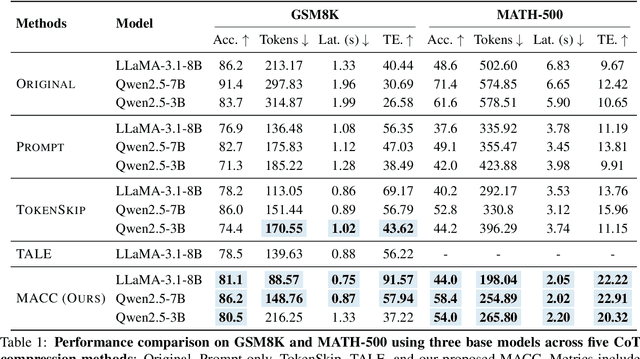
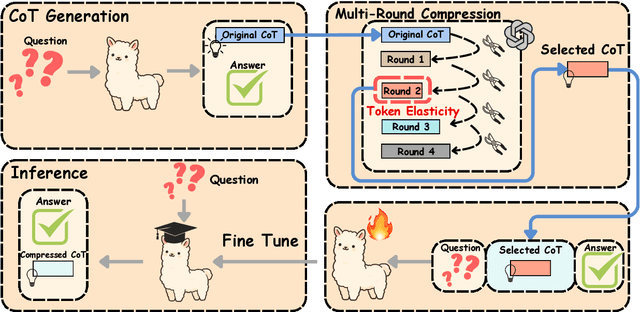
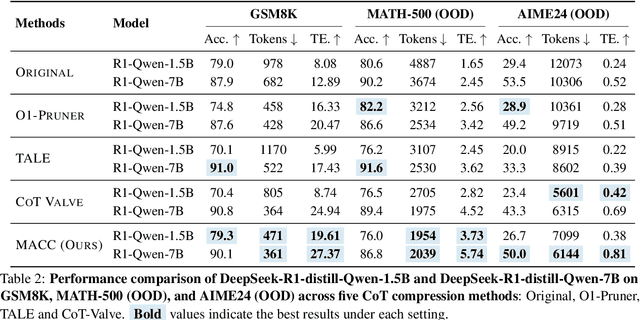
Abstract:Chain-of-Thought (CoT) reasoning improves performance on complex tasks but introduces significant inference latency due to verbosity. We propose Multiround Adaptive Chain-of-Thought Compression (MACC), a framework that leverages the token elasticity phenomenon--where overly small token budgets can paradoxically increase output length--to progressively compress CoTs via multiround refinement. This adaptive strategy allows MACC to determine the optimal compression depth for each input. Our method achieves an average accuracy improvement of 5.6 percent over state-of-the-art baselines, while also reducing CoT length by an average of 47 tokens and significantly lowering latency. Furthermore, we show that test-time performance--accuracy and token length--can be reliably predicted using interpretable features like perplexity and compression rate on the training set. Evaluated across different models, our method enables efficient model selection and forecasting without repeated fine-tuning, demonstrating that CoT compression is both effective and predictable. Our code will be released in https://github.com/Leon221220/MACC.
Learning in Order! A Sequential Strategy to Learn Invariant Features for Multimodal Sentiment Analysis
Sep 05, 2024



Abstract:This work proposes a novel and simple sequential learning strategy to train models on videos and texts for multimodal sentiment analysis. To estimate sentiment polarities on unseen out-of-distribution data, we introduce a multimodal model that is trained either in a single source domain or multiple source domains using our learning strategy. This strategy starts with learning domain invariant features from text, followed by learning sparse domain-agnostic features from videos, assisted by the selected features learned in text. Our experimental results demonstrate that our model achieves significantly better performance than the state-of-the-art approaches on average in both single-source and multi-source settings. Our feature selection procedure favors the features that are independent to each other and are strongly correlated with their polarity labels. To facilitate research on this topic, the source code of this work will be publicly available upon acceptance.
Advancing Biomedical Text Mining with Community Challenges
Mar 07, 2024Abstract:The field of biomedical research has witnessed a significant increase in the accumulation of vast amounts of textual data from various sources such as scientific literatures, electronic health records, clinical trial reports, and social media. However, manually processing and analyzing these extensive and complex resources is time-consuming and inefficient. To address this challenge, biomedical text mining, also known as biomedical natural language processing, has garnered great attention. Community challenge evaluation competitions have played an important role in promoting technology innovation and interdisciplinary collaboration in biomedical text mining research. These challenges provide platforms for researchers to develop state-of-the-art solutions for data mining and information processing in biomedical research. In this article, we review the recent advances in community challenges specific to Chinese biomedical text mining. Firstly, we collect the information of these evaluation tasks, such as data sources and task types. Secondly, we conduct systematic summary and comparative analysis, including named entity recognition, entity normalization, attribute extraction, relation extraction, event extraction, text classification, text similarity, knowledge graph construction, question answering, text generation, and large language model evaluation. Then, we summarize the potential clinical applications of these community challenge tasks from translational informatics perspective. Finally, we discuss the contributions and limitations of these community challenges, while highlighting future directions in the era of large language models.
Toward Robust Multimodal Learning using Multimodal Foundational Models
Jan 20, 2024Abstract:Existing multimodal sentiment analysis tasks are highly rely on the assumption that the training and test sets are complete multimodal data, while this assumption can be difficult to hold: the multimodal data are often incomplete in real-world scenarios. Therefore, a robust multimodal model in scenarios with randomly missing modalities is highly preferred. Recently, CLIP-based multimodal foundational models have demonstrated impressive performance on numerous multimodal tasks by learning the aligned cross-modal semantics of image and text pairs, but the multimodal foundational models are also unable to directly address scenarios involving modality absence. To alleviate this issue, we propose a simple and effective framework, namely TRML, Toward Robust Multimodal Learning using Multimodal Foundational Models. TRML employs generated virtual modalities to replace missing modalities, and aligns the semantic spaces between the generated and missing modalities. Concretely, we design a missing modality inference module to generate virtual modaliites and replace missing modalities. We also design a semantic matching learning module to align semantic spaces generated and missing modalities. Under the prompt of complete modality, our model captures the semantics of missing modalities by leveraging the aligned cross-modal semantic space. Experiments demonstrate the superiority of our approach on three multimodal sentiment analysis benchmark datasets, CMU-MOSI, CMU-MOSEI, and MELD.
Improving Natural Language Understanding with Computation-Efficient Retrieval Representation Fusion
Jan 04, 2024Abstract:Retrieval-based augmentations that aim to incorporate knowledge from an external database into language models have achieved great success in various knowledge-intensive (KI) tasks, such as question-answering and text generation. However, integrating retrievals in non-knowledge-intensive (NKI) tasks, such as text classification, is still challenging. Existing works focus on concatenating retrievals to inputs as context to form the prompt-based inputs. Unfortunately, such methods require language models to have the capability to handle long texts. Besides, inferring such concatenated data would also consume a significant amount of computational resources. To solve these challenges, we propose \textbf{ReFusion} in this paper, a computation-efficient \textbf{Re}trieval representation \textbf{Fusion} with neural architecture search. The main idea is to directly fuse the retrieval representations into the language models. Specifically, we first propose an online retrieval module that retrieves representations of similar sentences. Then, we present a retrieval fusion module including two effective ranking schemes, i.e., reranker-based scheme and ordered-mask-based scheme, to fuse the retrieval representations with hidden states. Furthermore, we use Neural Architecture Search (NAS) to seek the optimal fusion structure across different layers. Finally, we conduct comprehensive experiments, and the results demonstrate our ReFusion can achieve superior and robust performance on various NKI tasks.
Overview of the PromptCBLUE Shared Task in CHIP2023
Dec 29, 2023Abstract:This paper presents an overview of the PromptCBLUE shared task (http://cips-chip.org.cn/2023/eval1) held in the CHIP-2023 Conference. This shared task reformualtes the CBLUE benchmark, and provide a good testbed for Chinese open-domain or medical-domain large language models (LLMs) in general medical natural language processing. Two different tracks are held: (a) prompt tuning track, investigating the multitask prompt tuning of LLMs, (b) probing the in-context learning capabilities of open-sourced LLMs. Many teams from both the industry and academia participated in the shared tasks, and the top teams achieved amazing test results. This paper describes the tasks, the datasets, evaluation metrics, and the top systems for both tasks. Finally, the paper summarizes the techniques and results of the evaluation of the various approaches explored by the participating teams.
PromptCBLUE: A Chinese Prompt Tuning Benchmark for the Medical Domain
Oct 22, 2023



Abstract:Biomedical language understanding benchmarks are the driving forces for artificial intelligence applications with large language model (LLM) back-ends. However, most current benchmarks: (a) are limited to English which makes it challenging to replicate many of the successes in English for other languages, or (b) focus on knowledge probing of LLMs and neglect to evaluate how LLMs apply these knowledge to perform on a wide range of bio-medical tasks, or (c) have become a publicly available corpus and are leaked to LLMs during pre-training. To facilitate the research in medical LLMs, we re-build the Chinese Biomedical Language Understanding Evaluation (CBLUE) benchmark into a large scale prompt-tuning benchmark, PromptCBLUE. Our benchmark is a suitable test-bed and an online platform for evaluating Chinese LLMs' multi-task capabilities on a wide range bio-medical tasks including medical entity recognition, medical text classification, medical natural language inference, medical dialogue understanding and medical content/dialogue generation. To establish evaluation on these tasks, we have experimented and report the results with the current 9 Chinese LLMs fine-tuned with differtent fine-tuning techniques.
SHAPE: A Sample-adaptive Hierarchical Prediction Network for Medication Recommendation
Sep 09, 2023



Abstract:Effectively medication recommendation with complex multimorbidity conditions is a critical task in healthcare. Most existing works predicted medications based on longitudinal records, which assumed the information transmitted patterns of learning longitudinal sequence data are stable and intra-visit medical events are serialized. However, the following conditions may have been ignored: 1) A more compact encoder for intra-relationship in the intra-visit medical event is urgent; 2) Strategies for learning accurate representations of the variable longitudinal sequences of patients are different. In this paper, we proposed a novel Sample-adaptive Hierarchical medicAtion Prediction nEtwork, termed SHAPE, to tackle the above challenges in the medication recommendation task. Specifically, we design a compact intra-visit set encoder to encode the relationship in the medical event for obtaining visit-level representation and then develop an inter-visit longitudinal encoder to learn the patient-level longitudinal representation efficiently. To endow the model with the capability of modeling the variable visit length, we introduce a soft curriculum learning method to assign the difficulty of each sample automatically by the visit length. Extensive experiments on a benchmark dataset verify the superiority of our model compared with several state-of-the-art baselines.
Revisiting Event Argument Extraction: Can EAE Models Learn Better When Being Aware of Event Co-occurrences?
Jun 01, 2023



Abstract:Event co-occurrences have been proved effective for event extraction (EE) in previous studies, but have not been considered for event argument extraction (EAE) recently. In this paper, we try to fill this gap between EE research and EAE research, by highlighting the question that ``Can EAE models learn better when being aware of event co-occurrences?''. To answer this question, we reformulate EAE as a problem of table generation and extend a SOTA prompt-based EAE model into a non-autoregressive generation framework, called TabEAE, which is able to extract the arguments of multiple events in parallel. Under this framework, we experiment with 3 different training-inference schemes on 4 datasets (ACE05, RAMS, WikiEvents and MLEE) and discover that via training the model to extract all events in parallel, it can better distinguish the semantic boundary of each event and its ability to extract single event gets substantially improved. Experimental results show that our method achieves new state-of-the-art performance on the 4 datasets. Our code is avilable at https://github.com/Stardust-hyx/TabEAE.
CATNet: Cross-event Attention-based Time-aware Network for Medical Event Prediction
Apr 29, 2022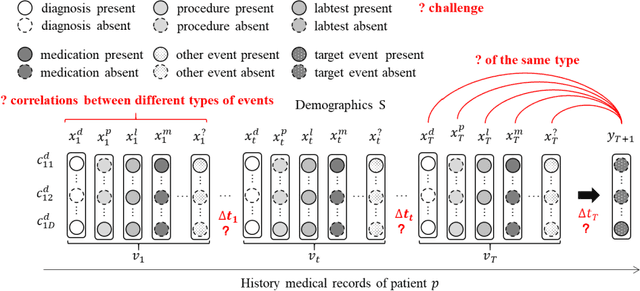

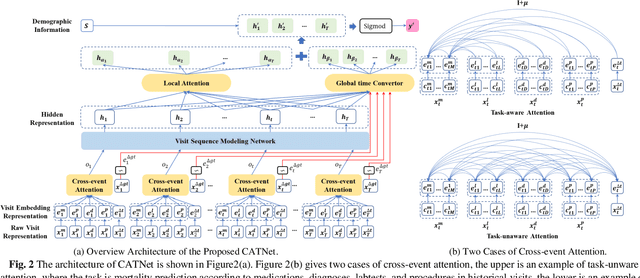
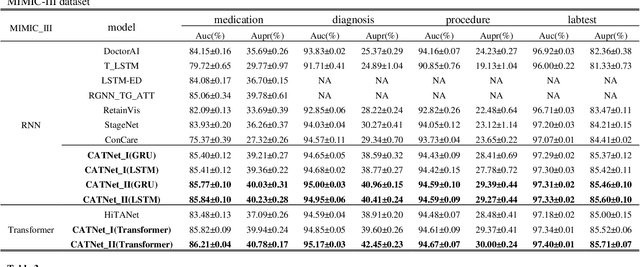
Abstract:Medical event prediction (MEP) is a fundamental task in the medical domain, which needs to predict medical events, including medications, diagnosis codes, laboratory tests, procedures, outcomes, and so on, according to historical medical records. The task is challenging as medical data is a type of complex time series data with heterogeneous and temporal irregular characteristics. Many machine learning methods that consider the two characteristics have been proposed for medical event prediction. However, most of them consider the two characteristics separately and ignore the correlations among different types of medical events, especially relations between historical medical events and target medical events. In this paper, we propose a novel neural network based on attention mechanism, called cross-event attention-based time-aware network (CATNet), for medical event prediction. It is a time-aware, event-aware and task-adaptive method with the following advantages: 1) modeling heterogeneous information and temporal information in a unified way and considering temporal irregular characteristics locally and globally respectively, 2) taking full advantage of correlations among different types of events via cross-event attention. Experiments on two public datasets (MIMIC-III and eICU) show CATNet can be adaptive with different MEP tasks and outperforms other state-of-the-art methods on various MEP tasks. The source code of CATNet will be released after this manuscript is accepted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge