Andrew P. King
King's College London
A Multimodal Deep Learning Model for Cardiac Resynchronisation Therapy Response Prediction
Jul 20, 2021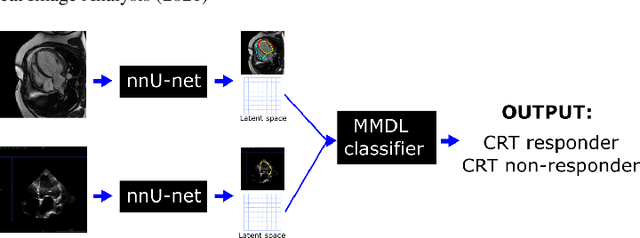
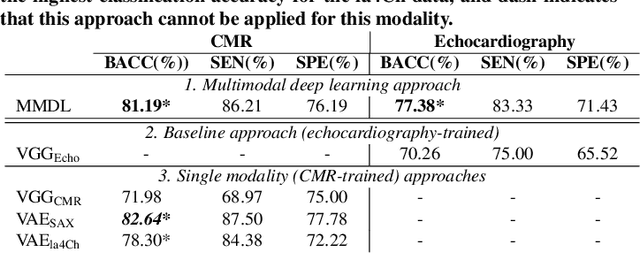
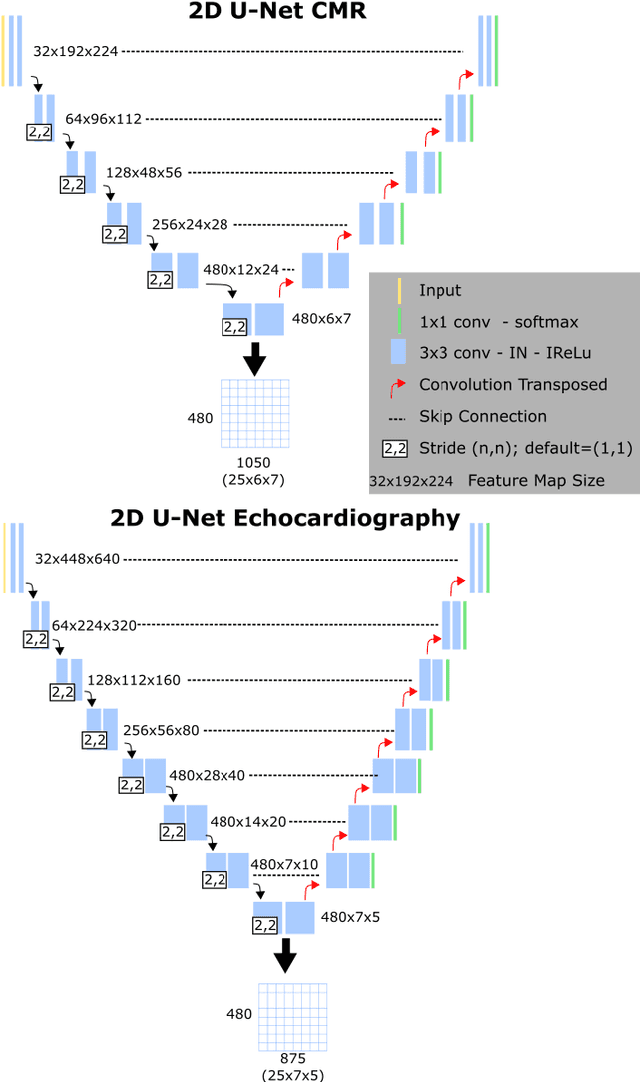
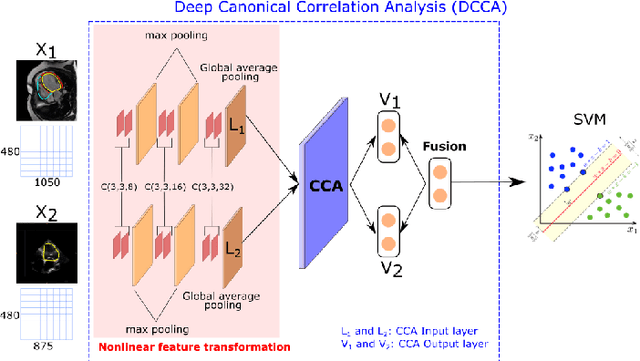
Abstract:We present a novel multimodal deep learning framework for cardiac resynchronisation therapy (CRT) response prediction from 2D echocardiography and cardiac magnetic resonance (CMR) data. The proposed method first uses the `nnU-Net' segmentation model to extract segmentations of the heart over the full cardiac cycle from the two modalities. Next, a multimodal deep learning classifier is used for CRT response prediction, which combines the latent spaces of the segmentation models of the two modalities. At inference time, this framework can be used with 2D echocardiography data only, whilst taking advantage of the implicit relationship between CMR and echocardiography features learnt from the model. We evaluate our pipeline on a cohort of 50 CRT patients for whom paired echocardiography/CMR data were available, and results show that the proposed multimodal classifier results in a statistically significant improvement in accuracy compared to the baseline approach that uses only 2D echocardiography data. The combination of multimodal data enables CRT response to be predicted with 77.38% accuracy (83.33% sensitivity and 71.43% specificity), which is comparable with the current state-of-the-art in machine learning-based CRT response prediction. Our work represents the first multimodal deep learning approach for CRT response prediction.
Fairness in Cardiac MR Image Analysis: An Investigation of Bias Due to Data Imbalance in Deep Learning Based Segmentation
Jul 01, 2021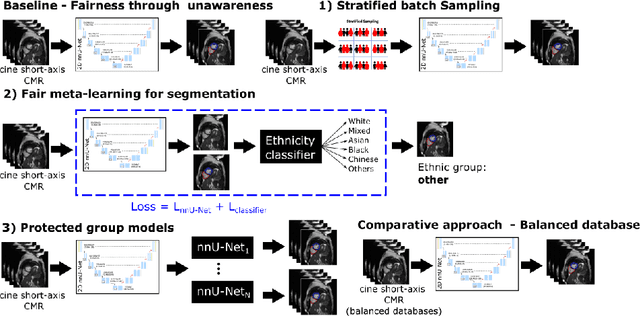
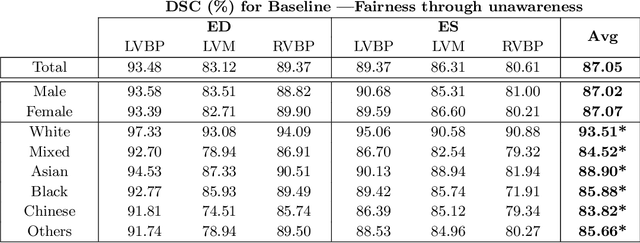
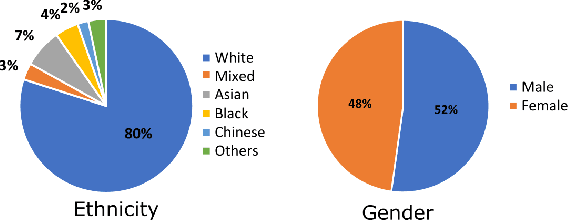
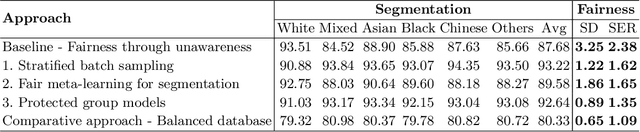
Abstract:The subject of "fairness" in artificial intelligence (AI) refers to assessing AI algorithms for potential bias based on demographic characteristics such as race and gender, and the development of algorithms to address this bias. Most applications to date have been in computer vision, although some work in healthcare has started to emerge. The use of deep learning (DL) in cardiac MR segmentation has led to impressive results in recent years, and such techniques are starting to be translated into clinical practice. However, no work has yet investigated the fairness of such models. In this work, we perform such an analysis for racial/gender groups, focusing on the problem of training data imbalance, using a nnU-Net model trained and evaluated on cine short axis cardiac MR data from the UK Biobank dataset, consisting of 5,903 subjects from 6 different racial groups. We find statistically significant differences in Dice performance between different racial groups. To reduce the racial bias, we investigated three strategies: (1) stratified batch sampling, in which batch sampling is stratified to ensure balance between racial groups; (2) fair meta-learning for segmentation, in which a DL classifier is trained to classify race and jointly optimized with the segmentation model; and (3) protected group models, in which a different segmentation model is trained for each racial group. We also compared the results to the scenario where we have a perfectly balanced database. To assess fairness we used the standard deviation (SD) and skewed error ratio (SER) of the average Dice values. Our results demonstrate that the racial bias results from the use of imbalanced training data, and that all proposed bias mitigation strategies improved fairness, with the best SD and SER resulting from the use of protected group models.
Channel Attention Networks for Robust MR Fingerprinting Matching
Dec 02, 2020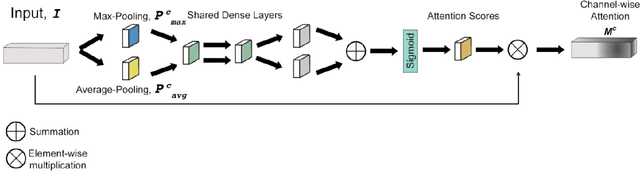
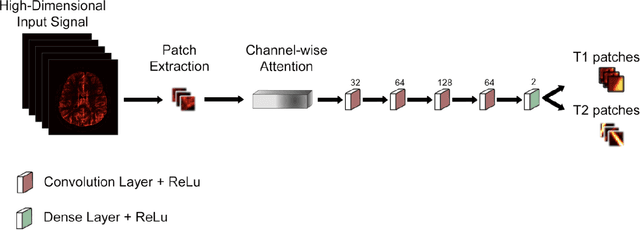
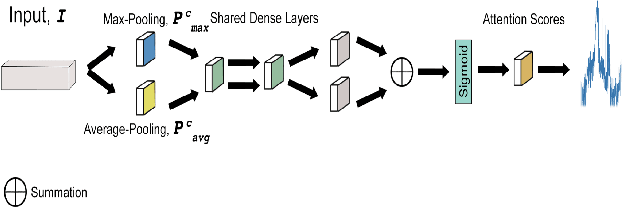
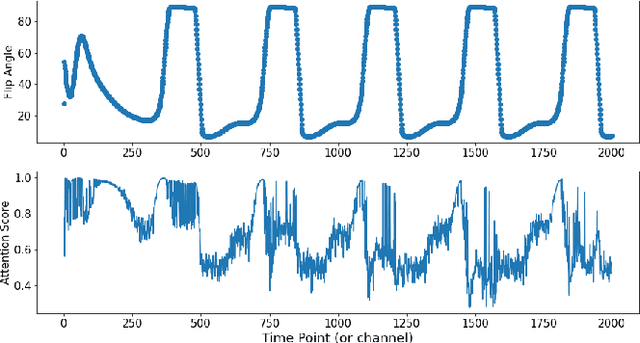
Abstract:Magnetic Resonance Fingerprinting (MRF) enables simultaneous mapping of multiple tissue parameters such as T1 and T2 relaxation times. The working principle of MRF relies on varying acquisition parameters pseudo-randomly, so that each tissue generates its unique signal evolution during scanning. Even though MRF provides faster scanning, it has disadvantages such as erroneous and slow generation of the corresponding parametric maps, which needs to be improved. Moreover, there is a need for explainable architectures for understanding the guiding signals to generate accurate parametric maps. In this paper, we addressed both of these shortcomings by proposing a novel neural network architecture consisting of a channel-wise attention module and a fully convolutional network. The proposed approach, evaluated over 3 simulated MRF signals, reduces error in the reconstruction of tissue parameters by 8.88% for T1 and 75.44% for T2 with respect to state-of-the-art methods. Another contribution of this study is a new channel selection method: attention-based channel selection. Furthermore, the effect of patch size and temporal frames of MRF signal on channel reduction are analyzed by employing a channel-wise attention.
Deep Learning for Automatic Spleen Length Measurement in Sickle Cell Disease Patients
Sep 06, 2020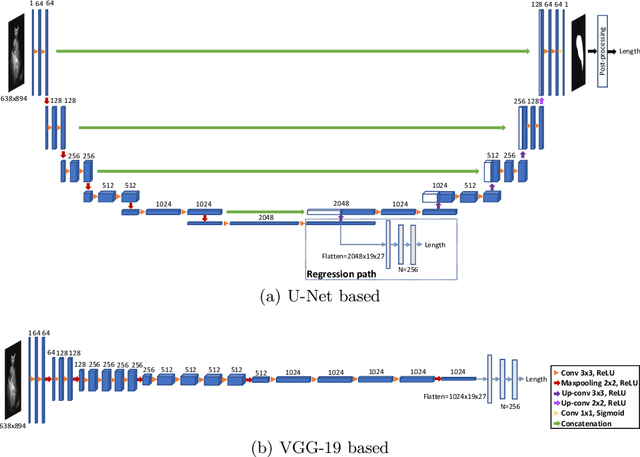
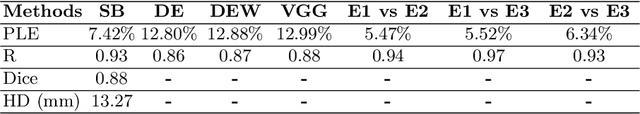
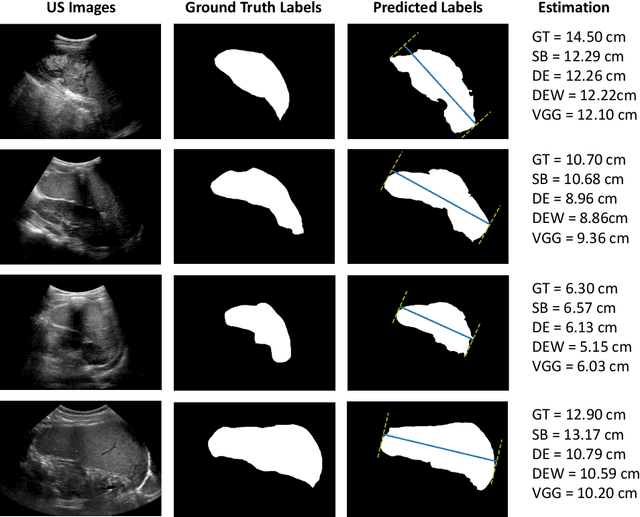
Abstract:Sickle Cell Disease (SCD) is one of the most common genetic diseases in the world. Splenomegaly (abnormal enlargement of the spleen) is frequent among children with SCD. If left untreated, splenomegaly can be life-threatening. The current workflow to measure spleen size includes palpation, possibly followed by manual length measurement in 2D ultrasound imaging. However, this manual measurement is dependent on operator expertise and is subject to intra- and inter-observer variability. We investigate the use of deep learning to perform automatic estimation of spleen length from ultrasound images. We investigate two types of approach, one segmentation-based and one based on direct length estimation, and compare the results against measurements made by human experts. Our best model (segmentation-based) achieved a percentage length error of 7.42%, which is approaching the level of inter-observer variability (5.47%-6.34%). To the best of our knowledge, this is the first attempt to measure spleen size in a fully automated way from ultrasound images.
Quality-aware semi-supervised learning for CMR segmentation
Sep 01, 2020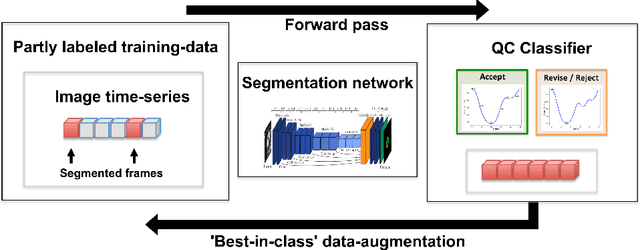
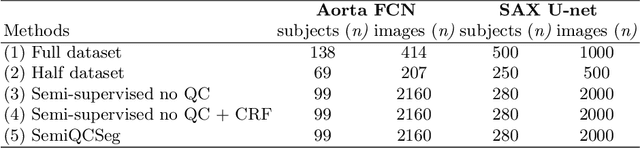
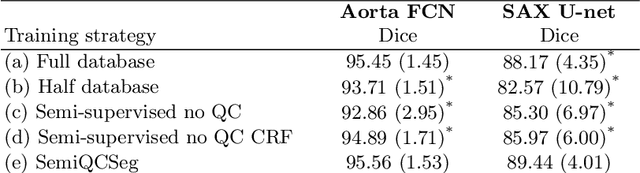
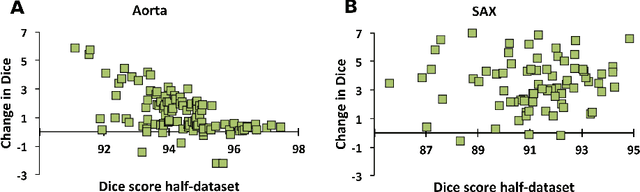
Abstract:One of the challenges in developing deep learning algorithms for medical image segmentation is the scarcity of annotated training data. To overcome this limitation, data augmentation and semi-supervised learning (SSL) methods have been developed. However, these methods have limited effectiveness as they either exploit the existing data set only (data augmentation) or risk negative impact by adding poor training examples (SSL). Segmentations are rarely the final product of medical image analysis - they are typically used in downstream tasks to infer higher-order patterns to evaluate diseases. Clinicians take into account a wealth of prior knowledge on biophysics and physiology when evaluating image analysis results. We have used these clinical assessments in previous works to create robust quality-control (QC) classifiers for automated cardiac magnetic resonance (CMR) analysis. In this paper, we propose a novel scheme that uses QC of the downstream task to identify high quality outputs of CMR segmentation networks, that are subsequently utilised for further network training. In essence, this provides quality-aware augmentation of training data in a variant of SSL for segmentation networks (semiQCSeg). We evaluate our approach in two CMR segmentation tasks (aortic and short axis cardiac volume segmentation) using UK Biobank data and two commonly used network architectures (U-net and a Fully Convolutional Network) and compare against supervised and SSL strategies. We show that semiQCSeg improves training of the segmentation networks. It decreases the need for labelled data, while outperforming the other methods in terms of Dice and clinical metrics. SemiQCSeg can be an efficient approach for training segmentation networks for medical image data when labelled datasets are scarce.
A persistent homology-based topological loss function for multi-class CNN segmentation of cardiac MRI
Aug 21, 2020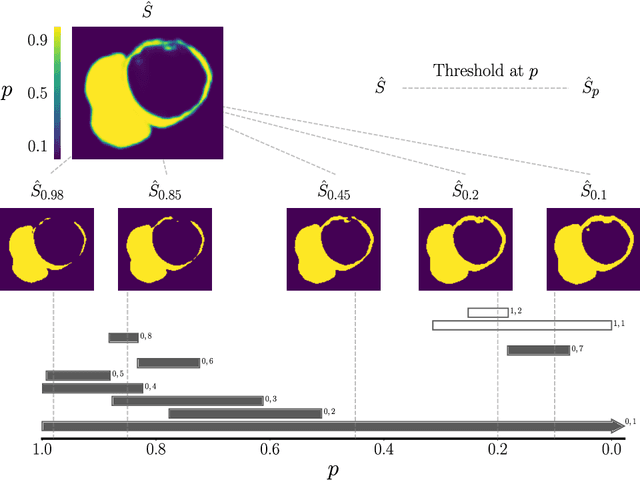
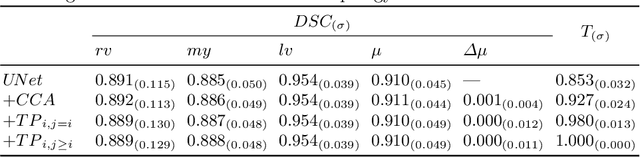
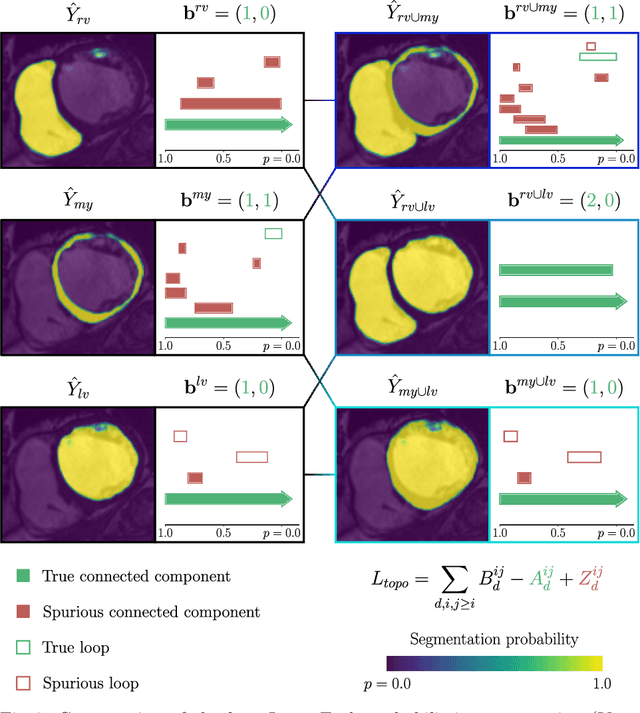
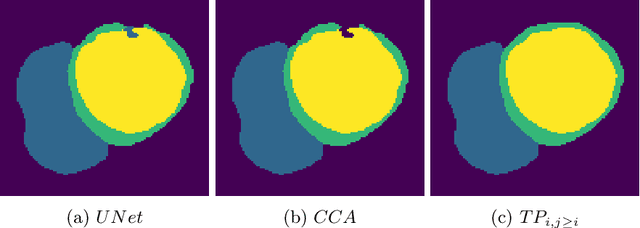
Abstract:With respect to spatial overlap, CNN-based segmentation of short axis cardiovascular magnetic resonance (CMR) images has achieved a level of performance consistent with inter observer variation. However, conventional training procedures frequently depend on pixel-wise loss functions, limiting optimisation with respect to extended or global features. As a result, inferred segmentations can lack spatial coherence, including spurious connected components or holes. Such results are implausible, violating the anticipated topology of image segments, which is frequently known a priori. Addressing this challenge, published work has employed persistent homology, constructing topological loss functions for the evaluation of image segments against an explicit prior. Building a richer description of segmentation topology by considering all possible labels and label pairs, we extend these losses to the task of multi-class segmentation. These topological priors allow us to resolve all topological errors in a subset of 150 examples from the ACDC short axis CMR training data set, without sacrificing overlap performance.
Interpretable Deep Models for Cardiac Resynchronisation Therapy Response Prediction
Jul 09, 2020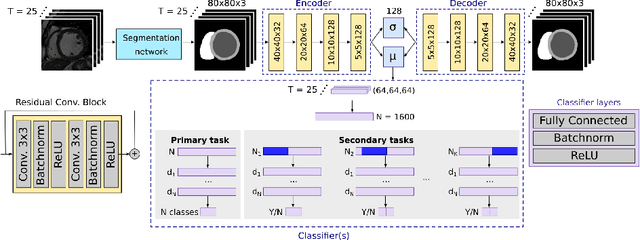
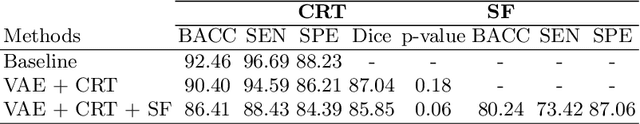
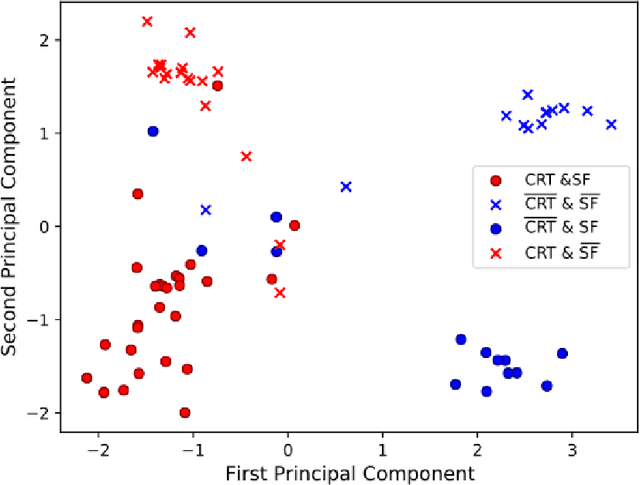

Abstract:Advances in deep learning (DL) have resulted in impressive accuracy in some medical image classification tasks, but often deep models lack interpretability. The ability of these models to explain their decisions is important for fostering clinical trust and facilitating clinical translation. Furthermore, for many problems in medicine there is a wealth of existing clinical knowledge to draw upon, which may be useful in generating explanations, but it is not obvious how this knowledge can be encoded into DL models - most models are learnt either from scratch or using transfer learning from a different domain. In this paper we address both of these issues. We propose a novel DL framework for image-based classification based on a variational autoencoder (VAE). The framework allows prediction of the output of interest from the latent space of the autoencoder, as well as visualisation (in the image domain) of the effects of crossing the decision boundary, thus enhancing the interpretability of the classifier. Our key contribution is that the VAE disentangles the latent space based on `explanations' drawn from existing clinical knowledge. The framework can predict outputs as well as explanations for these outputs, and also raises the possibility of discovering new biomarkers that are separate (or disentangled) from the existing knowledge. We demonstrate our framework on the problem of predicting response of patients with cardiomyopathy to cardiac resynchronization therapy (CRT) from cine cardiac magnetic resonance images. The sensitivity and specificity of the proposed model on the task of CRT response prediction are 88.43% and 84.39% respectively, and we showcase the potential of our model in enhancing understanding of the factors contributing to CRT response.
Automated quantification of myocardial tissue characteristics from native T1 mapping using neural networks with Bayesian inference for uncertainty-based quality-control
Jan 31, 2020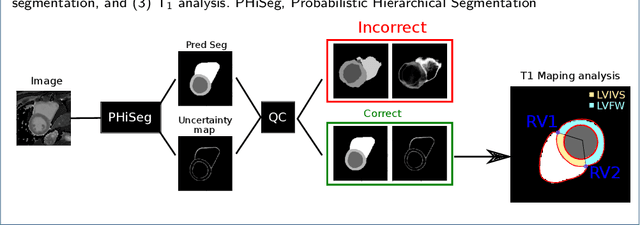
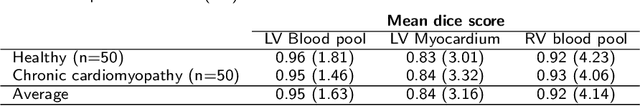
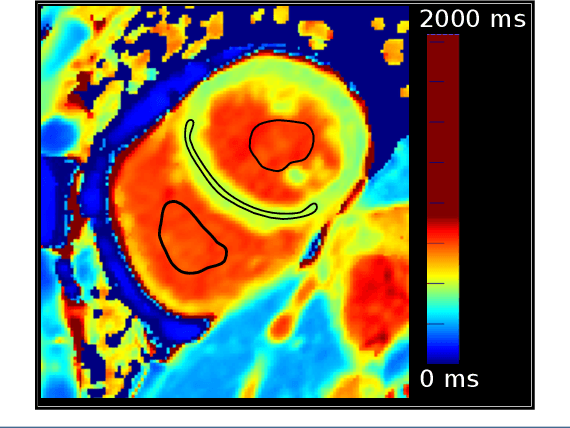

Abstract:Tissue characterisation with CMR parametric mapping has the potential to detect and quantify both focal and diffuse alterations in myocardial structure not assessable by late gadolinium enhancement. Native T1 mapping in particular has shown promise as a useful biomarker to support diagnostic, therapeutic and prognostic decision-making in ischaemic and non-ischaemic cardiomyopathies. Convolutional neural networks with Bayesian inference are a category of artificial neural networks which model the uncertainty of the network output. This study presents an automated framework for tissue characterisation from native ShMOLLI T1 mapping at 1.5T using a Probabilistic Hierarchical Segmentation (PHiSeg) network. In addition, we use the uncertainty information provided by the PHiSeg network in a novel automated quality control (QC) step to identify uncertain T1 values. The PHiSeg network and QC were validated against manual analysis on a cohort of the UK Biobank containing healthy subjects and chronic cardiomyopathy patients. We used the proposed method to obtain reference T1 ranges for the left ventricular myocardium in healthy subjects as well as common clinical cardiac conditions. T1 values computed from automatic and manual segmentations were highly correlated (r=0.97). Bland-Altman analysis showed good agreement between the automated and manual measurements. The average Dice metric was 0.84 for the left ventricular myocardium. The sensitivity of detection of erroneous outputs was 91%. Finally, T1 values were automatically derived from 14,683 CMR exams from the UK Biobank. The proposed pipeline allows for automatic analysis of myocardial native T1 mapping and includes a QC process to detect potentially erroneous results. T1 reference values were presented for healthy subjects and common clinical cardiac conditions from the largest cohort to date using T1-mapping images.
Deep Learning Based Detection and Correction of Cardiac MR Motion Artefacts During Reconstruction for High-Quality Segmentation
Oct 21, 2019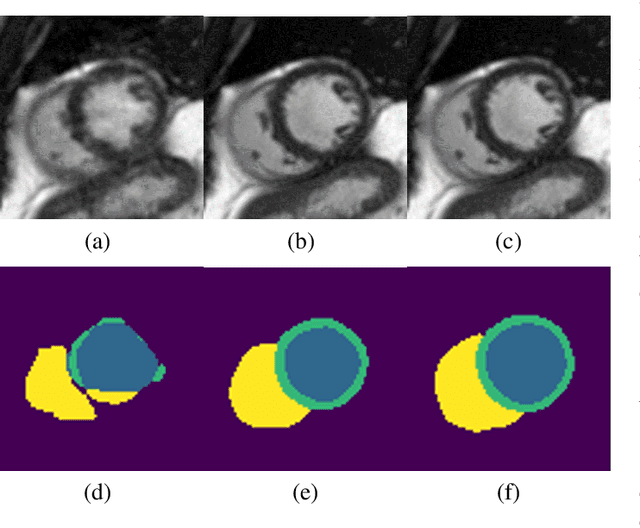
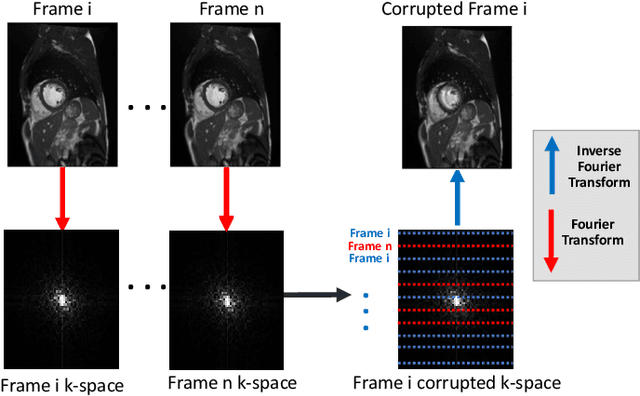
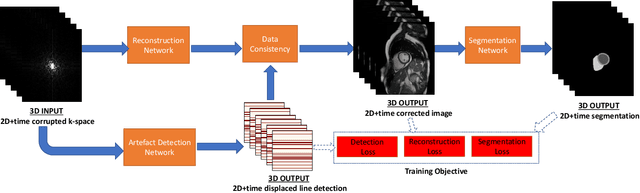
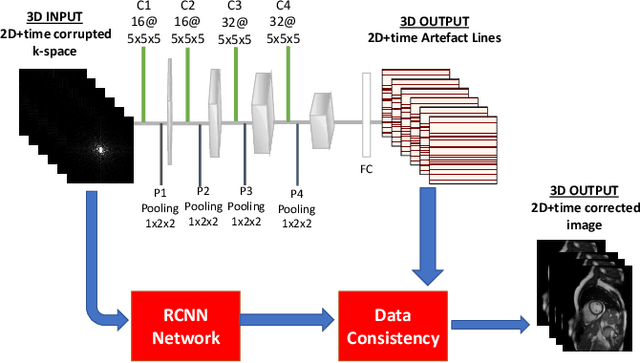
Abstract:Segmenting anatomical structures in medical images has been successfully addressed with deep learning methods for a range of applications. However, this success is heavily dependent on the quality of the image that is being segmented. A commonly neglected point in the medical image analysis community is the vast amount of clinical images that have severe image artefacts due to organ motion, movement of the patient and/or image acquisition related issues. In this paper, we discuss the implications of image motion artefacts on cardiac MR segmentation and compare a variety of approaches for jointly correcting for artefacts and segmenting the cardiac cavity. We propose to use a segmentation network coupled with this in an end-to-end framework. Our training optimises three different tasks: 1) image artefact detection, 2) artefact correction and 3) image segmentation. We train the reconstruction network to automatically correct for motion-related artefacts using synthetically corrupted cardiac MR k-space data and uncorrected reconstructed images. Using a test set of 500 2D+time cine MR acquisitions from the UK Biobank data set, we achieve demonstrably good image quality and high segmentation accuracy in the presence of synthetic motion artefacts. We quantitatively compare our method with a variety of techniques for jointly recovering image quality and performing image segmentation. We showcase better performance compared to state-of-the-art image correction techniques. Moreover, our method preserves the quality of uncorrupted images and therefore can be utilised as a global image reconstruction algorithm.
A Topological Loss Function for Deep-Learning based Image Segmentation using Persistent Homology
Oct 04, 2019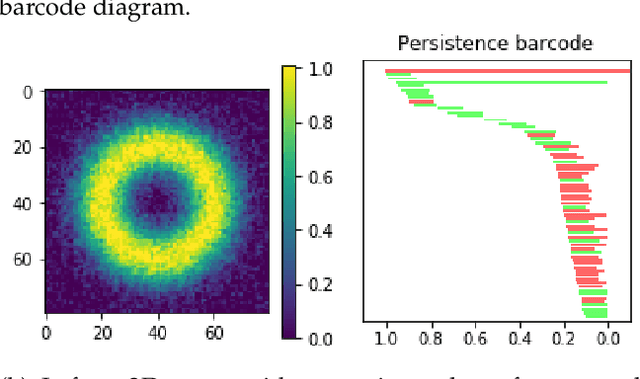
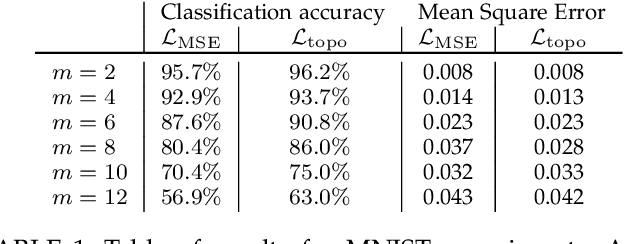
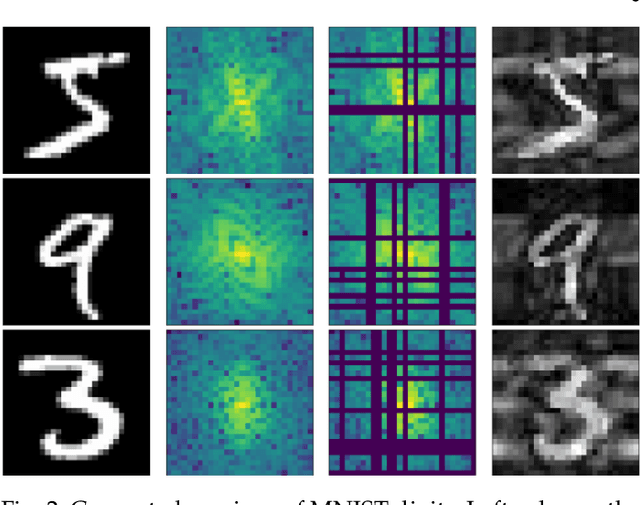
Abstract:We introduce a method for training neural networks to perform image or volume segmentation in which prior knowledge about the topology of the segmented object can be explicitly provided and then incorporated into the training process. By using the differentiable properties of persistent homology, a concept used in topological data analysis, we can specify the desired topology of segmented objects in terms of their Betti numbers and then drive the proposed segmentations to contain the specified topological features. Importantly this process does not require any ground-truth labels, just prior knowledge of the topology of the structure being segmented. We demonstrate our approach in three experiments. Firstly we create a synthetic task in which handwritten MNIST digits are de-noised, and show that using this kind of topological prior knowledge in the training of the network significantly improves the quality of the de-noised digits. Secondly we perform an experiment in which the task is segmenting the myocardium of the left ventricle from cardiac magnetic resonance images. We show that the incorporation of the prior knowledge of the topology of this anatomy improves the resulting segmentations in terms of both the topological accuracy and the Dice coefficient. Thirdly, we extend the method to 3D volumes and demonstrate its performance on the task of segmenting the placenta from ultrasound data, again showing that incorporating topological priors improves performance on this challenging task. We find that embedding explicit prior knowledge in neural network segmentation tasks is most beneficial when the segmentation task is especially challenging and that it can be used in either a semi-supervised or post-processing context to extract a useful training gradient from images without pixelwise labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge