Karthik Gopinath
JParc: Joint cortical surface parcellation with registration
Dec 27, 2025Abstract:Cortical surface parcellation is a fundamental task in both basic neuroscience research and clinical applications, enabling more accurate mapping of brain regions. Model-based and learning-based approaches for automated parcellation alleviate the need for manual labeling. Despite the advancement in parcellation performance, learning-based methods shift away from registration and atlas propagation without exploring the reason for the improvement compared to traditional methods. In this study, we present JParc, a joint cortical registration and parcellation framework, that outperforms existing state-of-the-art parcellation methods. In rigorous experiments, we demonstrate that the enhanced performance of JParc is primarily attributable to accurate cortical registration and a learned parcellation atlas. By leveraging a shallow subnetwork to fine-tune the propagated atlas labels, JParc achieves a Dice score greater than 90% on the Mindboggle dataset, using only basic geometric features (sulcal depth, curvature) that describe cortical folding patterns. The superior accuracy of JParc can significantly increase the statistical power in brain mapping studies as well as support applications in surgical planning and many other downstream neuroscientific and clinical tasks.
End-to-end Cortical Surface Reconstruction from Clinical Magnetic Resonance Images
May 20, 2025Abstract:Surface-based cortical analysis is valuable for a variety of neuroimaging tasks, such as spatial normalization, parcellation, and gray matter (GM) thickness estimation. However, most tools for estimating cortical surfaces work exclusively on scans with at least 1 mm isotropic resolution and are tuned to a specific magnetic resonance (MR) contrast, often T1-weighted (T1w). This precludes application using most clinical MR scans, which are very heterogeneous in terms of contrast and resolution. Here, we use synthetic domain-randomized data to train the first neural network for explicit estimation of cortical surfaces from scans of any contrast and resolution, without retraining. Our method deforms a template mesh to the white matter (WM) surface, which guarantees topological correctness. This mesh is further deformed to estimate the GM surface. We compare our method to recon-all-clinical (RAC), an implicit surface reconstruction method which is currently the only other tool capable of processing heterogeneous clinical MR scans, on ADNI and a large clinical dataset (n=1,332). We show a approximately 50 % reduction in cortical thickness error (from 0.50 to 0.24 mm) with respect to RAC and better recovery of the aging-related cortical thinning patterns detected by FreeSurfer on high-resolution T1w scans. Our method enables fast and accurate surface reconstruction of clinical scans, allowing studies (1) with sample sizes far beyond what is feasible in a research setting, and (2) of clinical populations that are difficult to enroll in research studies. The code is publicly available at https://github.com/simnibs/brainnet.
From Low Field to High Value: Robust Cortical Mapping from Low-Field MRI
May 18, 2025Abstract:Three-dimensional reconstruction of cortical surfaces from MRI for morphometric analysis is fundamental for understanding brain structure. While high-field MRI (HF-MRI) is standard in research and clinical settings, its limited availability hinders widespread use. Low-field MRI (LF-MRI), particularly portable systems, offers a cost-effective and accessible alternative. However, existing cortical surface analysis tools are optimized for high-resolution HF-MRI and struggle with the lower signal-to-noise ratio and resolution of LF-MRI. In this work, we present a machine learning method for 3D reconstruction and analysis of portable LF-MRI across a range of contrasts and resolutions. Our method works "out of the box" without retraining. It uses a 3D U-Net trained on synthetic LF-MRI to predict signed distance functions of cortical surfaces, followed by geometric processing to ensure topological accuracy. We evaluate our method using paired HF/LF-MRI scans of the same subjects, showing that LF-MRI surface reconstruction accuracy depends on acquisition parameters, including contrast type (T1 vs T2), orientation (axial vs isotropic), and resolution. A 3mm isotropic T2-weighted scan acquired in under 4 minutes, yields strong agreement with HF-derived surfaces: surface area correlates at r=0.96, cortical parcellations reach Dice=0.98, and gray matter volume achieves r=0.93. Cortical thickness remains more challenging with correlations up to r=0.70, reflecting the difficulty of sub-mm precision with 3mm voxels. We further validate our method on challenging postmortem LF-MRI, demonstrating its robustness. Our method represents a step toward enabling cortical surface analysis on portable LF-MRI. Code is available at https://surfer.nmr.mgh.harvard.edu/fswiki/ReconAny
Recon-all-clinical: Cortical surface reconstruction and analysis of heterogeneous clinical brain MRI
Sep 05, 2024



Abstract:Surface-based analysis of the cerebral cortex is ubiquitous in human neuroimaging with MRI. It is crucial for cortical registration, parcellation, and thickness estimation. Traditionally, these analyses require high-resolution, isotropic scans with good gray-white matter contrast, typically a 1mm T1-weighted scan. This excludes most clinical MRI scans, which are often anisotropic and lack the necessary T1 contrast. To enable large-scale neuroimaging studies using vast clinical data, we introduce recon-all-clinical, a novel method for cortical reconstruction, registration, parcellation, and thickness estimation in brain MRI scans of any resolution and contrast. Our approach employs a hybrid analysis method that combines a convolutional neural network (CNN) trained with domain randomization to predict signed distance functions (SDFs) and classical geometry processing for accurate surface placement while maintaining topological and geometric constraints. The method does not require retraining for different acquisitions, thus simplifying the analysis of heterogeneous clinical datasets. We tested recon-all-clinical on multiple datasets, including over 19,000 clinical scans. The method consistently produced precise cortical reconstructions and high parcellation accuracy across varied MRI contrasts and resolutions. Cortical thickness estimates are precise enough to capture aging effects independently of MRI contrast, although accuracy varies with slice thickness. Our method is publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical, enabling researchers to perform detailed cortical analysis on the huge amounts of already existing clinical MRI scans. This advancement may be particularly valuable for studying rare diseases and underrepresented populations where research-grade MRI data is scarce.
Registration by Regression (RbR): a framework for interpretable and flexible atlas registration
Apr 25, 2024


Abstract:In human neuroimaging studies, atlas registration enables mapping MRI scans to a common coordinate frame, which is necessary to aggregate data from multiple subjects. Machine learning registration methods have achieved excellent speed and accuracy but lack interpretability. More recently, keypoint-based methods have been proposed to tackle this issue, but their accuracy is still subpar, particularly when fitting nonlinear transforms. Here we propose Registration by Regression (RbR), a novel atlas registration framework that is highly robust and flexible, conceptually simple, and can be trained with cheaply obtained data. RbR predicts the (x,y,z) atlas coordinates for every voxel of the input scan (i.e., every voxel is a keypoint), and then uses closed-form expressions to quickly fit transforms using a wide array of possible deformation models, including affine and nonlinear (e.g., Bspline, Demons, invertible diffeomorphic models, etc.). Robustness is provided by the large number of voxels informing the registration and can be further increased by robust estimators like RANSAC. Experiments on independent public datasets show that RbR yields more accurate registration than competing keypoint approaches, while providing full control of the deformation model.
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies
May 02, 2023Abstract:Surface analysis of the cortex is ubiquitous in human neuroimaging with MRI, e.g., for cortical registration, parcellation, or thickness estimation. The convoluted cortical geometry requires isotropic scans (e.g., 1mm MPRAGEs) and good gray-white matter contrast for 3D reconstruction. This precludes the analysis of most brain MRI scans acquired for clinical purposes. Analyzing such scans would enable neuroimaging studies with sample sizes that cannot be achieved with current research datasets, particularly for underrepresented populations and rare diseases. Here we present the first method for cortical reconstruction, registration, parcellation, and thickness estimation for clinical brain MRI scans of any resolution and pulse sequence. The methods has a learning component and a classical optimization module. The former uses domain randomization to train a CNN that predicts an implicit representation of the white matter and pial surfaces (a signed distance function) at 1mm isotropic resolution, independently of the pulse sequence and resolution of the input. The latter uses geometry processing to place the surfaces while accurately satisfying topological and geometric constraints, thus enabling subsequent parcellation and thickness estimation with existing methods. We present results on 5mm axial FLAIR scans from ADNI and on a highly heterogeneous clinical dataset with 5,000 scans. Code and data are publicly available at https://surfer.nmr.mgh.harvard.edu/fswiki/recon-all-clinical
Graph Domain Adaptation for Alignment-Invariant Brain Surface Segmentation
Mar 31, 2020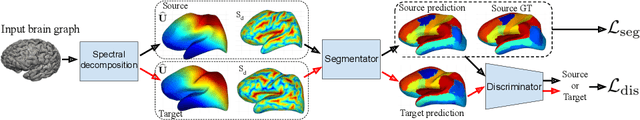
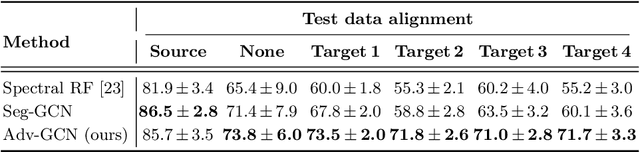
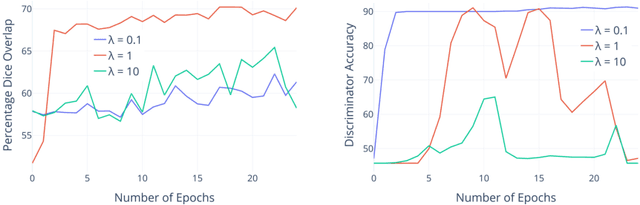
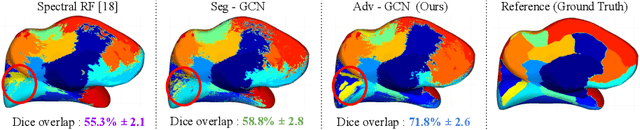
Abstract:The varying cortical geometry of the brain creates numerous challenges for its analysis. Recent developments have enabled learning surface data directly across multiple brain surfaces via graph convolutions on cortical data. However, current graph learning algorithms do fail when brain surface data are misaligned across subjects, thereby affecting their ability to deal with data from multiple domains. Adversarial training is widely used for domain adaptation to improve the segmentation performance across domains. In this paper, adversarial training is exploited to learn surface data across inconsistent graph alignments. This novel approach comprises a segmentator that uses a set of graph convolution layers to enable parcellation directly across brain surfaces in a source domain, and a discriminator that predicts a graph domain from segmentations. More precisely, the proposed adversarial network learns to generalize a parcellation across both, source and target domains. We demonstrate an 8% mean improvement in performance over a non-adversarial training strategy applied on multiple target domains extracted from MindBoggle, the largest publicly available manually-labeled brain surface dataset.
Learnable Pooling in Graph Convolution Networks for Brain Surface Analysis
Nov 22, 2019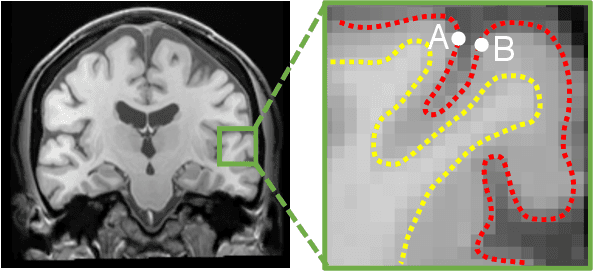
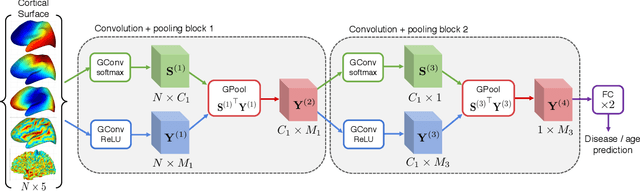
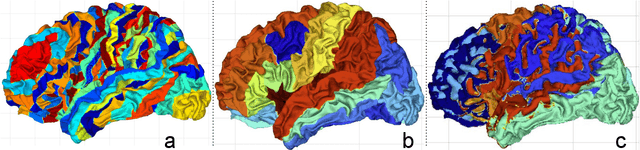
Abstract:Brain surface analysis is essential to neuroscience, however, the complex geometry of the brain cortex hinders computational methods for this task. The difficulty arises from a discrepancy between 3D imaging data, which is represented in Euclidean space, and the non-Euclidean geometry of the highly-convoluted brain surface. Recent advances in machine learning have enabled the use of neural networks for non-Euclidean spaces. These facilitate the learning of surface data, yet pooling strategies often remain constrained to a single fixed-graph. This paper proposes a new learnable graph pooling method for processing multiple surface-valued data to output subject-based information. The proposed method innovates by learning an intrinsic aggregation of graph nodes based on graph spectral embedding. We illustrate the advantages of our approach with in-depth experiments on two large-scale benchmark datasets. The flexibility of the pooling strategy is evaluated on four different prediction tasks, namely, subject-sex classification, regression of cortical region sizes, classification of Alzheimer's disease stages, and brain age regression. Our experiments demonstrate the superiority of our learnable pooling approach compared to other pooling techniques for graph convolution networks, with results improving the state-of-the-art in brain surface analysis.
Spectral Graph Transformer Networks for Brain Surface Parcellation
Nov 22, 2019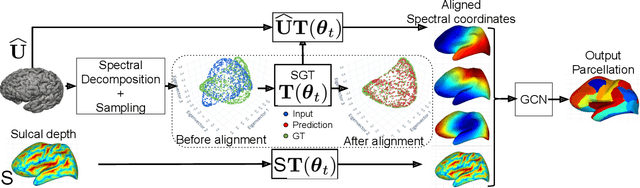
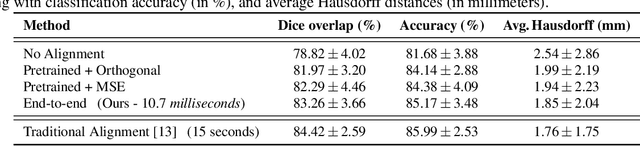
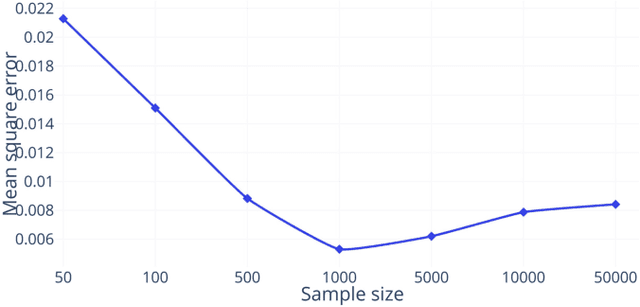
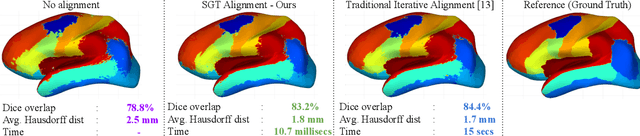
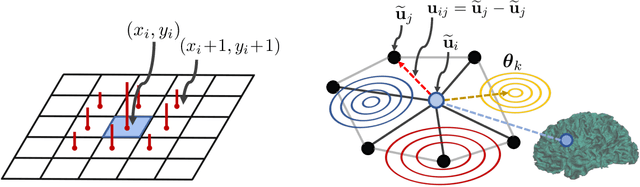
Abstract:The analysis of the brain surface modeled as a graph mesh is a challenging task. Conventional deep learning approaches often rely on data lying in the Euclidean space. As an extension to irregular graphs, convolution operations are defined in the Fourier or spectral domain. This spectral domain is obtained by decomposing the graph Laplacian, which captures relevant shape information. However, the spectral decomposition across different brain graphs causes inconsistencies between the eigenvectors of individual spectral domains, causing the graph learning algorithm to fail. Current spectral graph convolution methods handle this variance by separately aligning the eigenvectors to a reference brain in a slow iterative step. This paper presents a novel approach for learning the transformation matrix required for aligning brain meshes using a direct data-driven approach. Our alignment and graph processing method provides a fast analysis of brain surfaces. The novel Spectral Graph Transformer (SGT) network proposed in this paper uses very few randomly sub-sampled nodes in the spectral domain to learn the alignment matrix for multiple brain surfaces. We validate the use of this SGT network along with a graph convolution network to perform cortical parcellation. Our method on 101 manually-labeled brain surfaces shows improved parcellation performance over a no-alignment strategy, gaining a significant speed (1400 fold) over traditional iterative alignment approaches.
A deep learning framework for segmentation of retinal layers from OCT images
Jun 22, 2018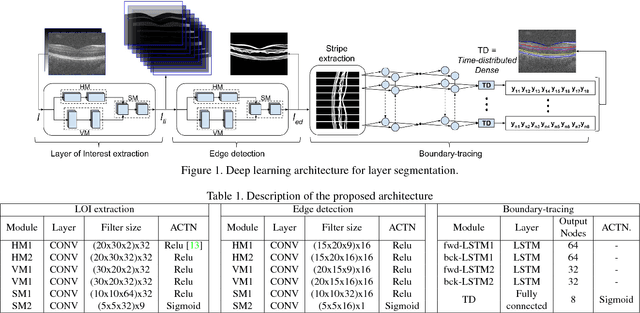
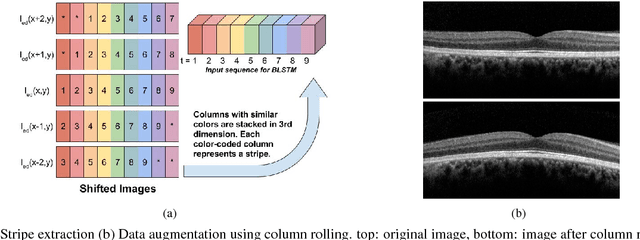
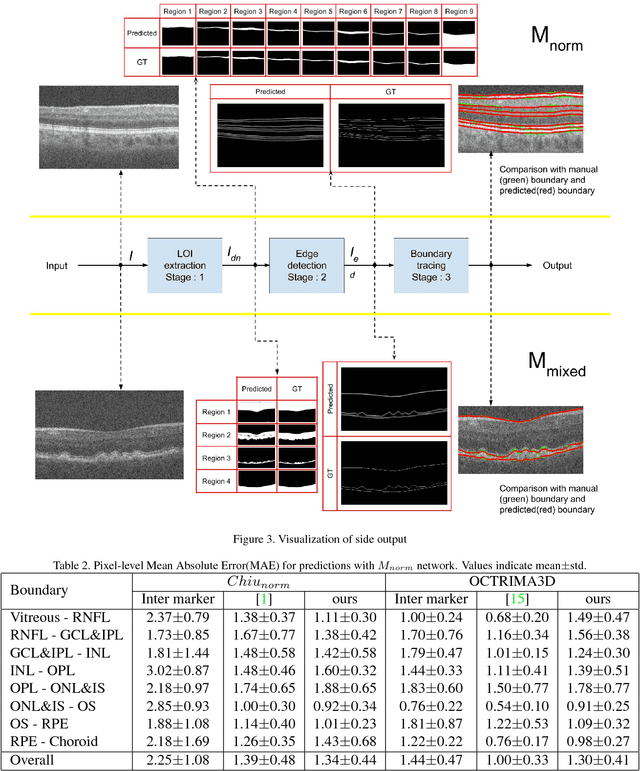
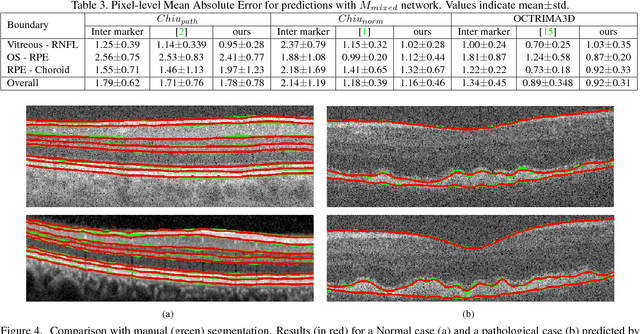
Abstract:Segmentation of retinal layers from Optical Coherence Tomography (OCT) volumes is a fundamental problem for any computer aided diagnostic algorithm development. This requires preprocessing steps such as denoising, region of interest extraction, flattening and edge detection all of which involve separate parameter tuning. In this paper, we explore deep learning techniques to automate all these steps and handle the presence/absence of pathologies. A model is proposed consisting of a combination of Convolutional Neural Network (CNN) and Long Short Term Memory (LSTM). The CNN is used to extract layers of interest image and extract the edges, while the LSTM is used to trace the layer boundary. This model is trained on a mixture of normal and AMD cases using minimal data. Validation results on three public datasets show that the pixel-wise mean absolute error obtained with our system is 1.30 plus or minus 0.48 which is lower than the inter-marker error of 1.79 plus or minus 0.76. Our model's performance is also on par with the existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge