Amirhossein Bayat
Department of Diagnostic and Interventional Neuroradiology, School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Germany, Department of Informatics, Technical University of Munich, Germany
A Computed Tomography Vertebral Segmentation Dataset with Anatomical Variations and Multi-Vendor Scanner Data
Mar 10, 2021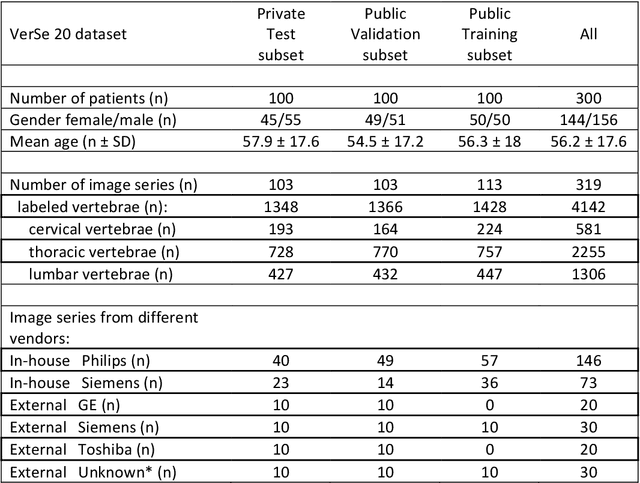
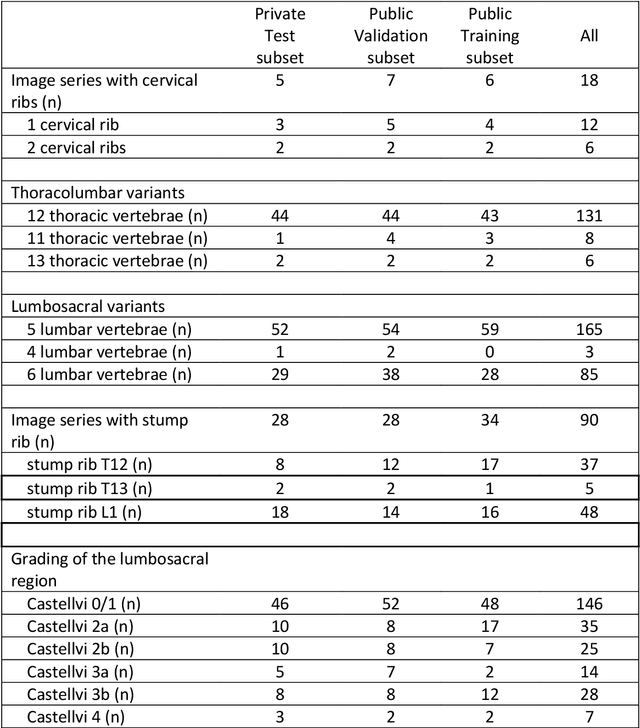
Abstract:With the advent of deep learning algorithms, fully automated radiological image analysis is within reach. In spine imaging, several atlas- and shape-based as well as deep learning segmentation algorithms have been proposed, allowing for subsequent automated analysis of morphology and pathology. The first Large Scale Vertebrae Segmentation Challenge (VerSe 2019) showed that these perform well on normal anatomy, but fail in variants not frequently present in the training dataset. Building on that experience, we report on the largely increased VerSe 2020 dataset and results from the second iteration of the VerSe challenge (MICCAI 2020, Lima, Peru). VerSe 2020 comprises annotated spine computed tomography (CT) images from 300 subjects with 4142 fully visualized and annotated vertebrae, collected across multiple centres from four different scanner manufacturers, enriched with cases that exhibit anatomical variants such as enumeration abnormalities (n=77) and transitional vertebrae (n=161). Metadata includes vertebral labelling information, voxel-level segmentation masks obtained with a human-machine hybrid algorithm and anatomical ratings, to enable the development and benchmarking of robust and accurate segmentation algorithms.
Federated Multi-Mini-Batch: An Efficient Training Approach to Federated Learning in Non-IID Environments
Nov 13, 2020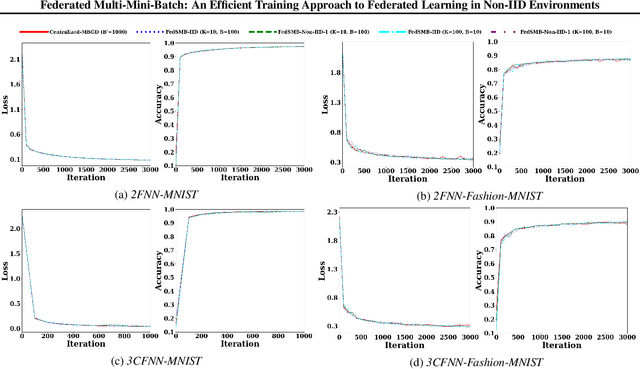
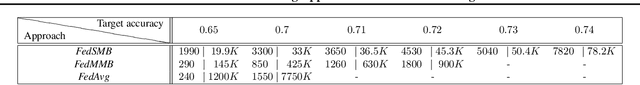
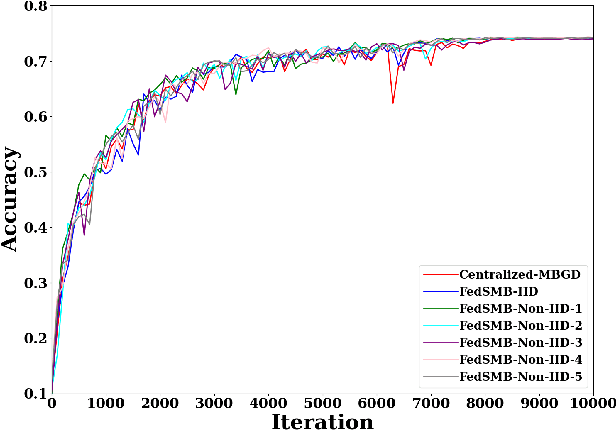
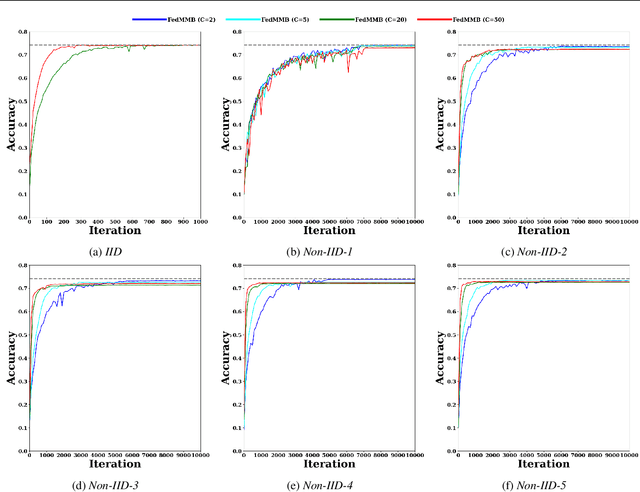
Abstract:Federated learning is a well-established approach to privacy-preserving training of a joint model on heavily distributed data. Federated averaging (FedAvg) is a well-known communication-efficient algorithm for federated learning, which performs well if the data distribution across the clients is independently and identically distributed (IID). However, FedAvg provides a lower accuracy and still requires a large number of communication rounds to achieve a target accuracy when it comes to Non-IID environments. To address the former limitation, we present federated single mini-batch (FedSMB), where the clients train the model on a single mini-batch from their dataset in each iteration. We show that FedSMB achieves the accuracy of the centralized training in Non-IID configurations, but in a considerable number of iterations. To address the latter limitation, we introduce federated multi-mini-batch (FedMMB) as a generalization of FedSMB, where the clients train the model on multiple mini-batches (specified by the batch count) in each communication round. FedMMB decouples the batch size from the batch count and provides a trade-off between the accuracy and communication efficiency in Non-IID settings. This is not possible with FedAvg, in which a single parameter determines both the batch size and batch count. The simulation results illustrate that FedMMB outperforms FedAvg in terms of the accuracy, communication efficiency, as well as computational efficiency and is an efficient training approach to federated learning in Non-IID environments.
Cranial Implant Prediction using Low-Resolution 3D Shape Completion and High-Resolution 2D Refinement
Sep 27, 2020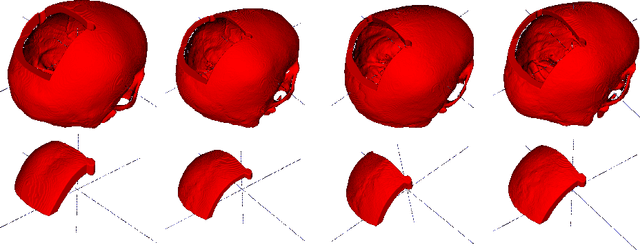
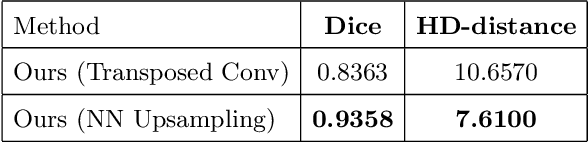
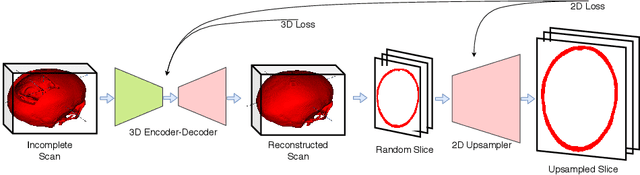
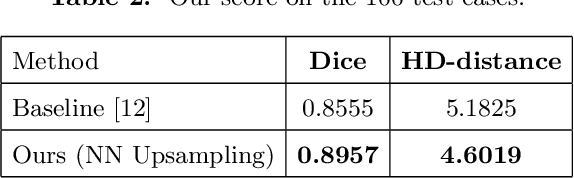
Abstract:Designing of a cranial implant needs a 3D understanding of the complete skull shape. Thus, taking a 2D approach is sub-optimal, since a 2D model lacks a holistic 3D view of both the defective and healthy skulls. Further, loading the whole 3D skull shapes at its original image resolution is not feasible in commonly available GPUs. To mitigate these issues, we propose a fully convolutional network composed of two subnetworks. The first subnetwork is designed to complete the shape of the downsampled defective skull. The second subnetwork upsamples the reconstructed shape slice-wise. We train the 3D and 2D networks together end-to-end, with a hierarchical loss function. Our proposed solution accurately predicts a high-resolution 3D implant in the challenge test case in terms of dice-score and the Hausdorff distance.
Robustification of Segmentation Models Against Adversarial Perturbations In Medical Imaging
Sep 23, 2020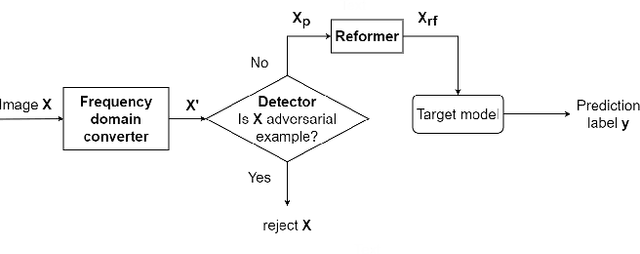
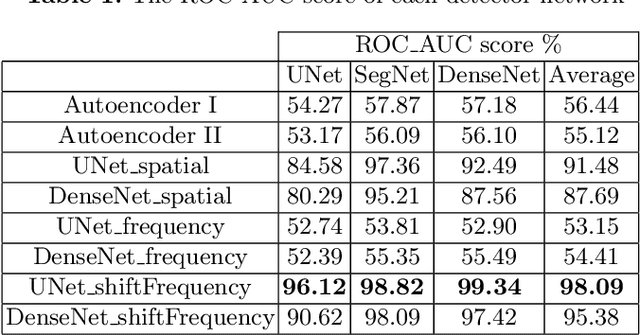
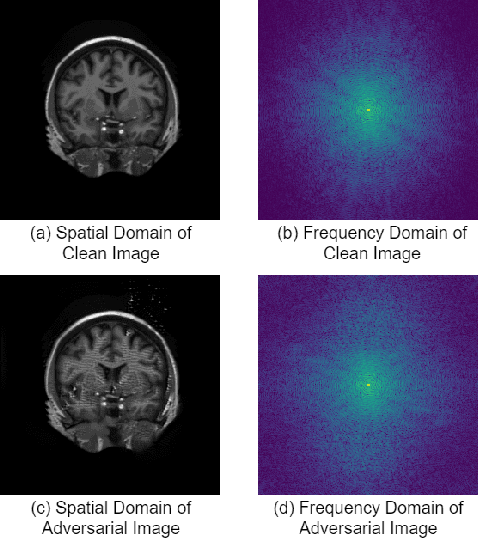
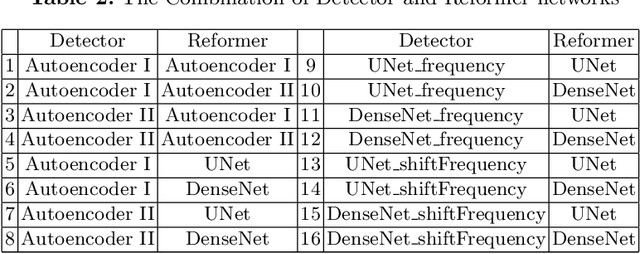
Abstract:This paper presents a novel yet efficient defense framework for segmentation models against adversarial attacks in medical imaging. In contrary to the defense methods against adversarial attacks for classification models which widely are investigated, such defense methods for segmentation models has been less explored. Our proposed method can be used for any deep learning models without revising the target deep learning models, as well as can be independent of adversarial attacks. Our framework consists of a frequency domain converter, a detector, and a reformer. The frequency domain converter helps the detector detects adversarial examples by using a frame domain of an image. The reformer helps target models to predict more precisely. We have experiments to empirically show that our proposed method has a better performance compared to the existing defense method.
Inferring the 3D Standing Spine Posture from 2D Radiographs
Jul 13, 2020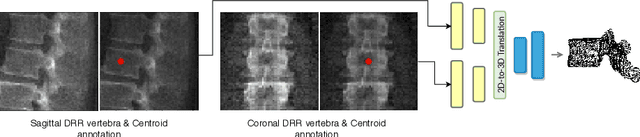
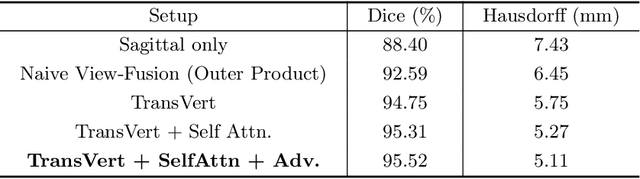
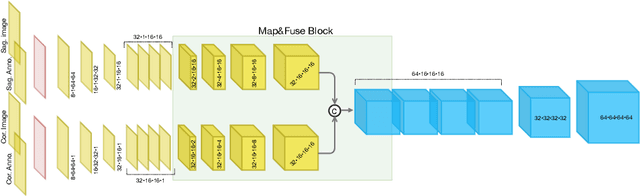
Abstract:The treatment of degenerative spinal disorders requires an understanding of the individual spinal anatomy and curvature in 3D. An upright spinal pose (i.e. standing) under natural weight bearing is crucial for such bio-mechanical analysis. 3D volumetric imaging modalities (e.g. CT and MRI) are performed in patients lying down. On the other hand, radiographs are captured in an upright pose, but result in 2D projections. This work aims to integrate the two realms, i.e. it combines the upright spinal curvature from radiographs with the 3D vertebral shape from CT imaging for synthesizing an upright 3D model of spine, loaded naturally. Specifically, we propose a novel neural network architecture working vertebra-wise, termed \emph{TransVert}, which takes orthogonal 2D radiographs and infers the spine's 3D posture. We validate our architecture on digitally reconstructed radiographs, achieving a 3D reconstruction Dice of $95.52\%$, indicating an almost perfect 2D-to-3D domain translation. Deploying our model on clinical radiographs, we successfully synthesise full-3D, upright, patient-specific spine models for the first time.
A distance-based loss for smooth and continuous skin layer segmentation in optoacoustic images
Jul 10, 2020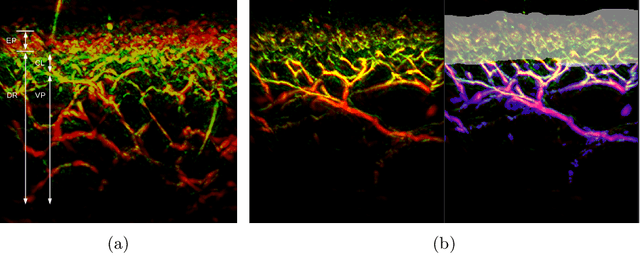
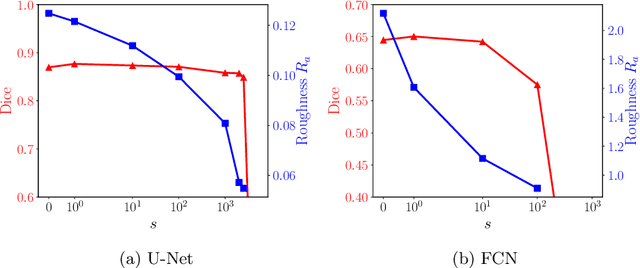

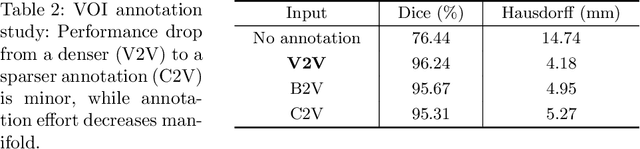
Abstract:Raster-scan optoacoustic mesoscopy (RSOM) is a powerful, non-invasive optical imaging technique for functional, anatomical, and molecular skin and tissue analysis. However, both the manual and the automated analysis of such images are challenging, because the RSOM images have very low contrast, poor signal to noise ratio, and systematic overlaps between the absorption spectra of melanin and hemoglobin. Nonetheless, the segmentation of the epidermis layer is a crucial step for many downstream medical and diagnostic tasks, such as vessel segmentation or monitoring of cancer progression. We propose a novel, shape-specific loss function that overcomes discontinuous segmentations and achieves smooth segmentation surfaces while preserving the same volumetric Dice and IoU. Further, we validate our epidermis segmentation through the sensitivity of vessel segmentation. We found a 20 $\%$ improvement in Dice for vessel segmentation tasks when the epidermis mask is provided as additional information to the vessel segmentation network.
Feedback Graph Attention Convolutional Network for Medical Image Enhancement
Jun 24, 2020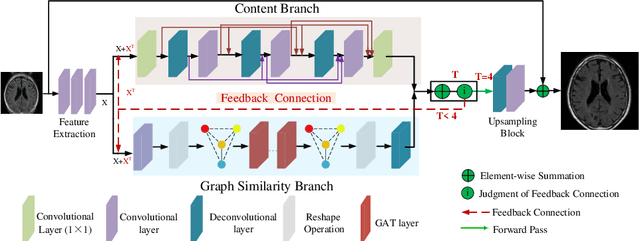
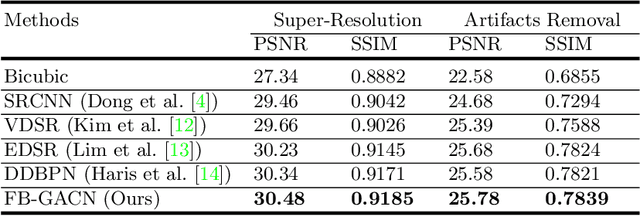
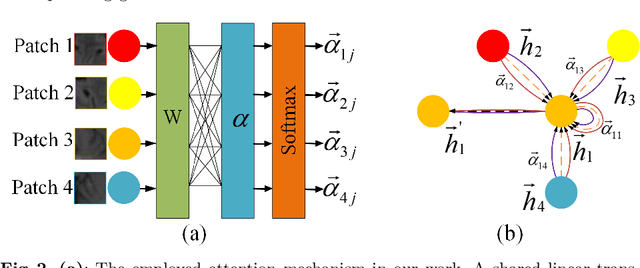

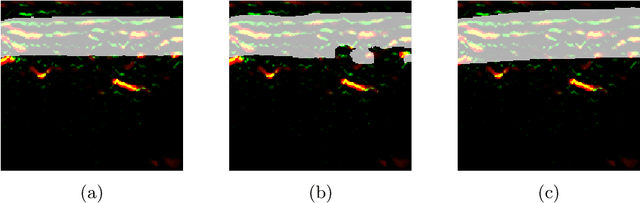
Abstract:Artifacts, blur and noise are the common distortions degrading MRI images during the acquisition process, and deep neural networks have been demonstrated to help in improving image quality. To well exploit global structural information and texture details, we propose a novel biomedical image enhancement network, named Feedback Graph Attention Convolutional Network (FB-GACN). As a key innovation, we consider the global structure of an image by building a graph network from image sub-regions that we consider to be node features, linking them non-locally according to their similarity. The proposed model consists of three main parts: 1) The parallel graph similarity branch and content branch, where the graph similarity branch aims at exploiting the similarity and symmetry across different image sub-regions in low-resolution feature space and provides additional priors for the content branch to enhance texture details. 2) A feedback mechanism with a recurrent structure to refine low-level representations with high-level information and generate powerful high-level texture details by handling the feedback connections. 3) A reconstruction to remove the artifacts and recover super-resolution images by using the estimated sub-region correlation priors obtained from the graph similarity branch. We evaluate our method on two image enhancement tasks: i) cross-protocol super resolution of diffusion MRI; ii) artifact removal of FLAIR MR images. Experimental results demonstrate that the proposed algorithm outperforms the state-of-the-art methods.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020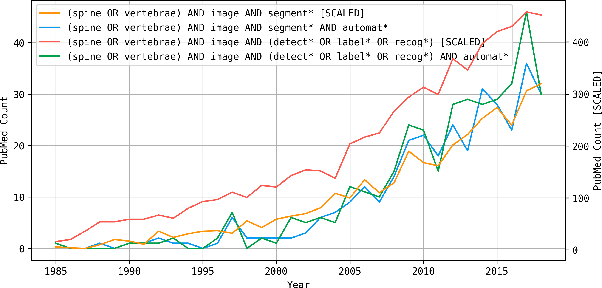
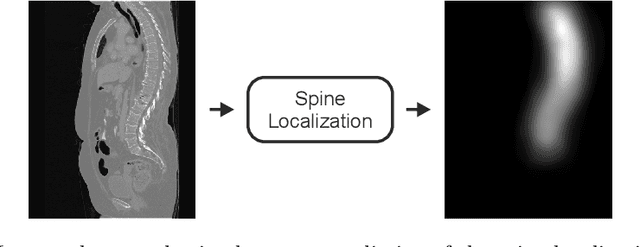
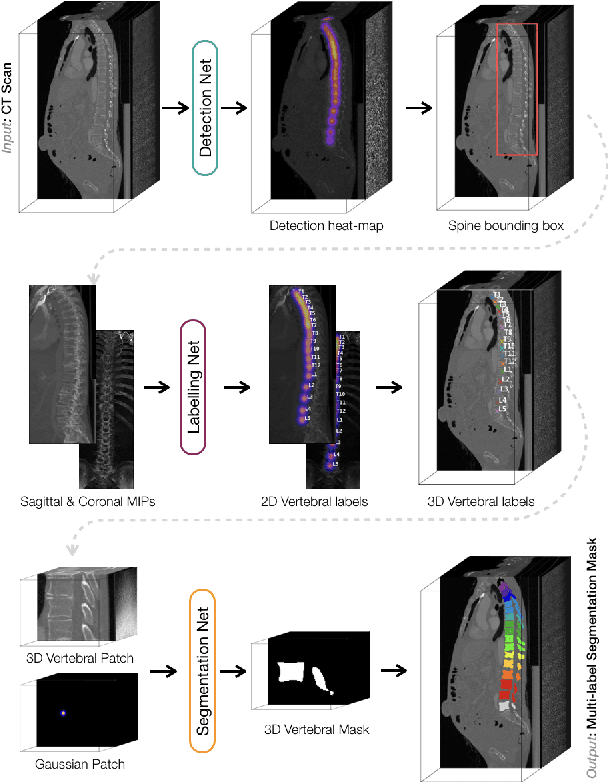
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge