Darko Stern
Augmentation-based Domain Generalization and Joint Training from Multiple Source Domains for Whole Heart Segmentation
Aug 06, 2025Abstract:As the leading cause of death worldwide, cardiovascular diseases motivate the development of more sophisticated methods to analyze the heart and its substructures from medical images like Computed Tomography (CT) and Magnetic Resonance (MR). Semantic segmentations of important cardiac structures that represent the whole heart are useful to assess patient-specific cardiac morphology and pathology. Furthermore, accurate semantic segmentations can be used to generate cardiac digital twin models which allows e.g. electrophysiological simulation and personalized therapy planning. Even though deep learning-based methods for medical image segmentation achieved great advancements over the last decade, retaining good performance under domain shift -- i.e. when training and test data are sampled from different data distributions -- remains challenging. In order to perform well on domains known at training-time, we employ a (1) balanced joint training approach that utilizes CT and MR data in equal amounts from different source domains. Further, aiming to alleviate domain shift towards domains only encountered at test-time, we rely on (2) strong intensity and spatial augmentation techniques to greatly diversify the available training data. Our proposed whole heart segmentation method, a 5-fold ensemble with our contributions, achieves the best performance for MR data overall and a performance similar to the best performance for CT data when compared to a model trained solely on CT. With 93.33% DSC and 0.8388 mm ASSD for CT and 89.30% DSC and 1.2411 mm ASSD for MR data, our method demonstrates great potential to efficiently obtain accurate semantic segmentations from which patient-specific cardiac twin models can be generated.
LA-CaRe-CNN: Cascading Refinement CNN for Left Atrial Scar Segmentation
Aug 06, 2025Abstract:Atrial fibrillation (AF) represents the most prevalent type of cardiac arrhythmia for which treatment may require patients to undergo ablation therapy. In this surgery cardiac tissues are locally scarred on purpose to prevent electrical signals from causing arrhythmia. Patient-specific cardiac digital twin models show great potential for personalized ablation therapy, however, they demand accurate semantic segmentation of healthy and scarred tissue typically obtained from late gadolinium enhanced (LGE) magnetic resonance (MR) scans. In this work we propose the Left Atrial Cascading Refinement CNN (LA-CaRe-CNN), which aims to accurately segment the left atrium as well as left atrial scar tissue from LGE MR scans. LA-CaRe-CNN is a 2-stage CNN cascade that is trained end-to-end in 3D, where Stage 1 generates a prediction for the left atrium, which is then refined in Stage 2 in conjunction with the original image information to obtain a prediction for the left atrial scar tissue. To account for domain shift towards domains unknown during training, we employ strong intensity and spatial augmentation to increase the diversity of the training dataset. Our proposed method based on a 5-fold ensemble achieves great segmentation results, namely, 89.21% DSC and 1.6969 mm ASSD for the left atrium, as well as 64.59% DSC and 91.80% G-DSC for the more challenging left atrial scar tissue. Thus, segmentations obtained through LA-CaRe-CNN show great potential for the generation of patient-specific cardiac digital twin models and downstream tasks like personalized targeted ablation therapy to treat AF.
Teeth Localization and Lesion Segmentation in CBCT Images using SpatialConfiguration-Net and U-Net
Dec 19, 2023Abstract:The localization of teeth and segmentation of periapical lesions in cone-beam computed tomography (CBCT) images are crucial tasks for clinical diagnosis and treatment planning, which are often time-consuming and require a high level of expertise. However, automating these tasks is challenging due to variations in shape, size, and orientation of lesions, as well as similar topologies among teeth. Moreover, the small volumes occupied by lesions in CBCT images pose a class imbalance problem that needs to be addressed. In this study, we propose a deep learning-based method utilizing two convolutional neural networks: the SpatialConfiguration-Net (SCN) and a modified version of the U-Net. The SCN accurately predicts the coordinates of all teeth present in an image, enabling precise cropping of teeth volumes that are then fed into the U-Net which detects lesions via segmentation. To address class imbalance, we compare the performance of three reweighting loss functions. After evaluation on 144 CBCT images, our method achieves a 97.3% accuracy for teeth localization, along with a promising sensitivity and specificity of 0.97 and 0.88, respectively, for subsequent lesion detection.
Efficient Multi-Organ Segmentation Using SpatialConfiguration-Net with Low GPU Memory Requirements
Nov 26, 2021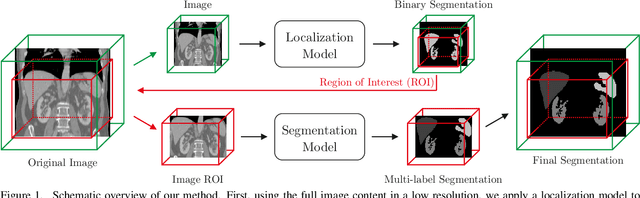

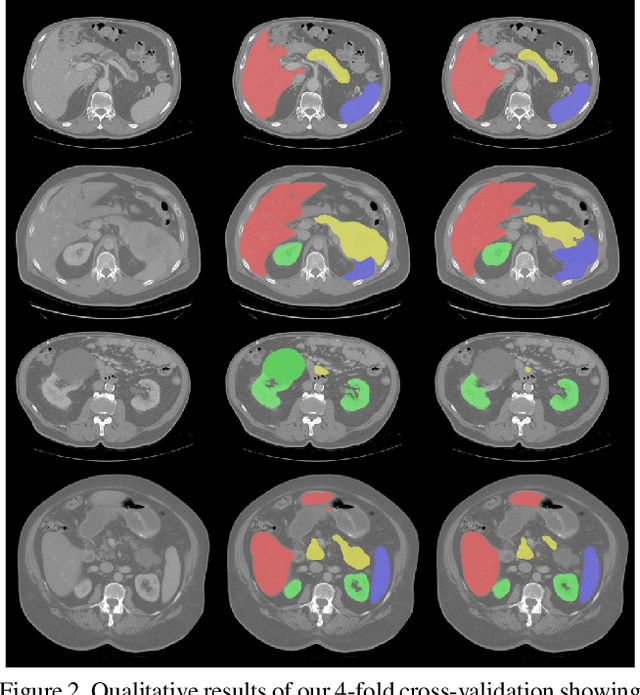
Abstract:Even though many semantic segmentation methods exist that are able to perform well on many medical datasets, often, they are not designed for direct use in clinical practice. The two main concerns are generalization to unseen data with a different visual appearance, e.g., images acquired using a different scanner, and efficiency in terms of computation time and required Graphics Processing Unit (GPU) memory. In this work, we employ a multi-organ segmentation model based on the SpatialConfiguration-Net (SCN), which integrates prior knowledge of the spatial configuration among the labelled organs to resolve spurious responses in the network outputs. Furthermore, we modified the architecture of the segmentation model to reduce its memory footprint as much as possible without drastically impacting the quality of the predictions. Lastly, we implemented a minimal inference script for which we optimized both, execution time and required GPU memory.
Modeling Annotation Uncertainty with Gaussian Heatmaps in Landmark Localization
Sep 21, 2021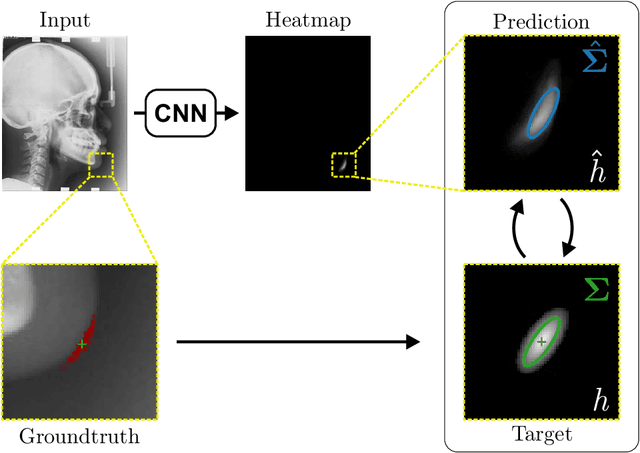
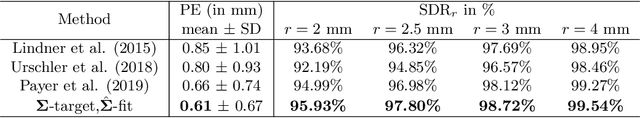
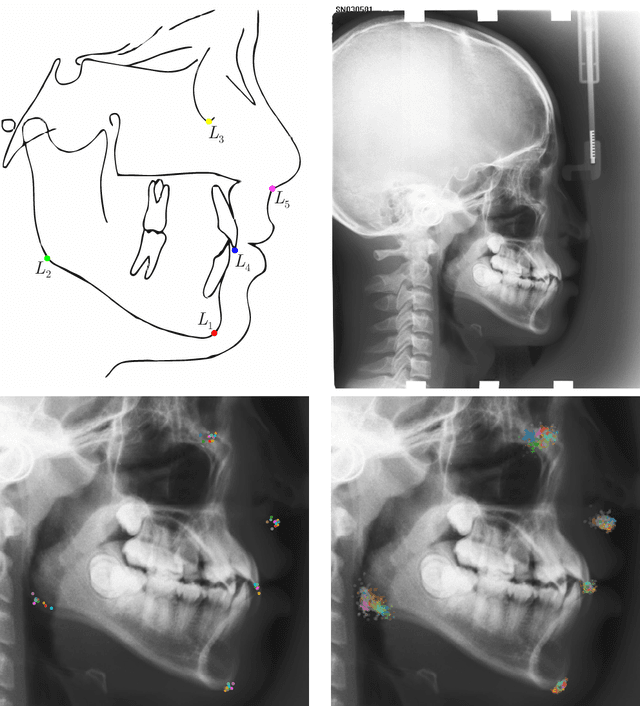
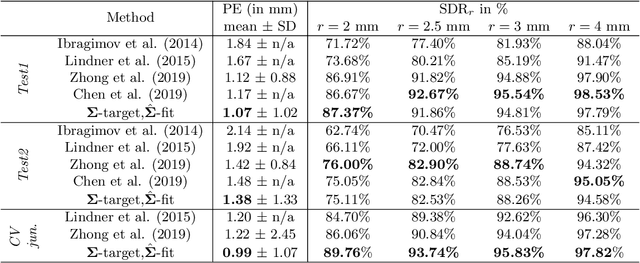
Abstract:In landmark localization, due to ambiguities in defining their exact position, landmark annotations may suffer from large observer variabilities, which result in uncertain annotations. To model the annotation ambiguities of the training dataset, we propose to learn anisotropic Gaussian parameters modeling the shape of the target heatmap during optimization. Furthermore, our method models the prediction uncertainty of individual samples by fitting anisotropic Gaussian functions to the predicted heatmaps during inference. Besides state-of-the-art results, our experiments on datasets of hand radiographs and lateral cephalograms also show that Gaussian functions are correlated with both localization accuracy and observer variability. As a final experiment, we show the importance of integrating the uncertainty into decision making by measuring the influence of the predicted location uncertainty on the classification of anatomical abnormalities in lateral cephalograms.
Inferring the 3D Standing Spine Posture from 2D Radiographs
Jul 13, 2020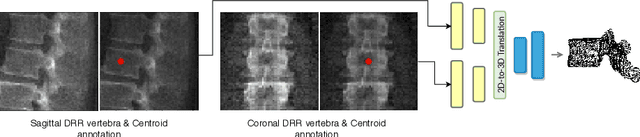
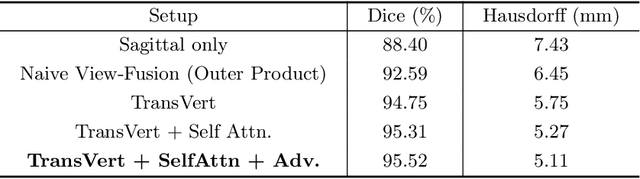
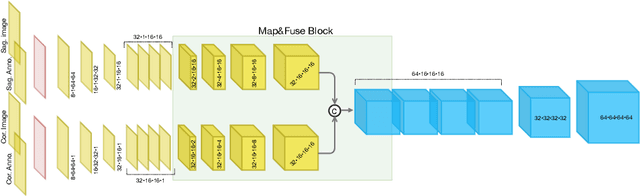
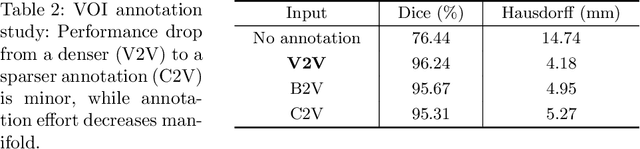
Abstract:The treatment of degenerative spinal disorders requires an understanding of the individual spinal anatomy and curvature in 3D. An upright spinal pose (i.e. standing) under natural weight bearing is crucial for such bio-mechanical analysis. 3D volumetric imaging modalities (e.g. CT and MRI) are performed in patients lying down. On the other hand, radiographs are captured in an upright pose, but result in 2D projections. This work aims to integrate the two realms, i.e. it combines the upright spinal curvature from radiographs with the 3D vertebral shape from CT imaging for synthesizing an upright 3D model of spine, loaded naturally. Specifically, we propose a novel neural network architecture working vertebra-wise, termed \emph{TransVert}, which takes orthogonal 2D radiographs and infers the spine's 3D posture. We validate our architecture on digitally reconstructed radiographs, achieving a 3D reconstruction Dice of $95.52\%$, indicating an almost perfect 2D-to-3D domain translation. Deploying our model on clinical radiographs, we successfully synthesise full-3D, upright, patient-specific spine models for the first time.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019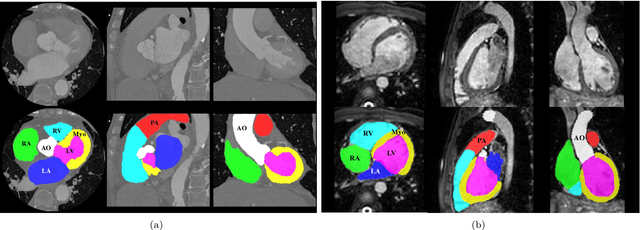
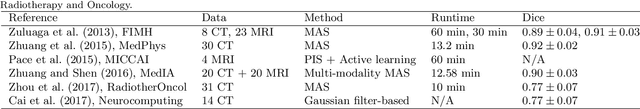
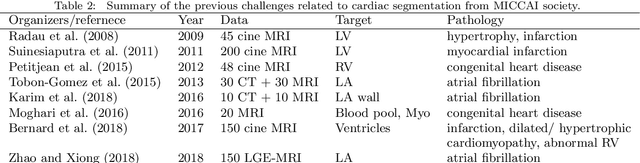
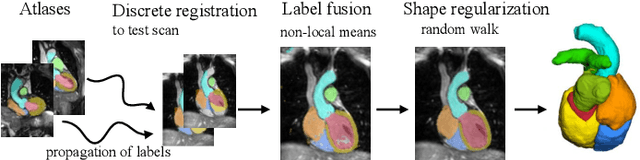
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge