Ahmed Abdulkadir
from the iSTAGING consortium, for the ADNI
Learning Actionable World Models for Industrial Process Control
Mar 03, 2025



Abstract:To go from (passive) process monitoring to active process control, an effective AI system must learn about the behavior of the complex system from very limited training data, forming an ad-hoc digital twin with respect to process in- and outputs that captures the consequences of actions on the process's world. We propose a novel methodology based on learning world models that disentangles process parameters in the learned latent representation, allowing for fine-grained control. Representation learning is driven by the latent factors that influence the processes through contrastive learning within a joint embedding predictive architecture. This makes changes in representations predictable from changes in inputs and vice versa, facilitating interpretability of key factors responsible for process variations, paving the way for effective control actions to keep the process within operational bounds. The effectiveness of our method is validated on the example of plastic injection molding, demonstrating practical relevance in proposing specific control actions for a notoriously unstable process.
AI Agents for Computer Use: A Review of Instruction-based Computer Control, GUI Automation, and Operator Assistants
Jan 27, 2025



Abstract:Instruction-based computer control agents (CCAs) execute complex action sequences on personal computers or mobile devices to fulfill tasks using the same graphical user interfaces as a human user would, provided instructions in natural language. This review offers a comprehensive overview of the emerging field of instruction-based computer control, examining available agents -- their taxonomy, development, and respective resources -- and emphasizing the shift from manually designed, specialized agents to leveraging foundation models such as large language models (LLMs) and vision-language models (VLMs). We formalize the problem and establish a taxonomy of the field to analyze agents from three perspectives: (a) the environment perspective, analyzing computer environments; (b) the interaction perspective, describing observations spaces (e.g., screenshots, HTML) and action spaces (e.g., mouse and keyboard actions, executable code); and (c) the agent perspective, focusing on the core principle of how an agent acts and learns to act. Our framework encompasses both specialized and foundation agents, facilitating their comparative analysis and revealing how prior solutions in specialized agents, such as an environment learning step, can guide the development of more capable foundation agents. Additionally, we review current CCA datasets and CCA evaluation methods and outline the challenges to deploying such agents in a productive setting. In total, we review and classify 86 CCAs and 33 related datasets. By highlighting trends, limitations, and future research directions, this work presents a comprehensive foundation to obtain a broad understanding of the field and push its future development.
A Comprehensive Survey of Deep Transfer Learning for Anomaly Detection in Industrial Time Series: Methods, Applications, and Directions
Jul 11, 2023Abstract:Automating the monitoring of industrial processes has the potential to enhance efficiency and optimize quality by promptly detecting abnormal events and thus facilitating timely interventions. Deep learning, with its capacity to discern non-trivial patterns within large datasets, plays a pivotal role in this process. Standard deep learning methods are suitable to solve a specific task given a specific type of data. During training, the algorithms demand large volumes of labeled training data. However, due to the dynamic nature of processes and the environment, it is impractical to acquire the needed data for standard deep learning training for every slightly different case anew. Deep transfer learning offers a solution to this problem. By leveraging knowledge from related tasks and accounting for variations in data distributions, this learning framework solves new tasks even with little or no additional labeled data. The approach bypasses the need to retrain a model from scratch for every new setup and dramatically reduces the labeled data requirement. This survey provides an in-depth review of deep transfer learning, examining the problem settings of transfer learning and classifying the prevailing deep transfer learning methods. Moreover, we delve into applying deep transfer learning in the context of a broad spectrum of time series anomaly detection tasks prevalent in primary industrial domains, e.g., manufacturing process monitoring, predictive maintenance, energy management, and infrastructure facility monitoring. We conclude this survey by underlining the challenges and limitations of deep transfer learning in industrial contexts. We also provide practical directions for solution design and implementation for these tasks, leading to specific, actionable suggestions.
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023


Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Explainable, Domain-Adaptive, and Federated Artificial Intelligence in Medicine
Nov 17, 2022



Abstract:Artificial intelligence (AI) continues to transform data analysis in many domains. Progress in each domain is driven by a growing body of annotated data, increased computational resources, and technological innovations. In medicine, the sensitivity of the data, the complexity of the tasks, the potentially high stakes, and a requirement of accountability give rise to a particular set of challenges. In this review, we focus on three key methodological approaches that address some of the particular challenges in AI-driven medical decision making. (1) Explainable AI aims to produce a human-interpretable justification for each output. Such models increase confidence if the results appear plausible and match the clinicians expectations. However, the absence of a plausible explanation does not imply an inaccurate model. Especially in highly non-linear, complex models that are tuned to maximize accuracy, such interpretable representations only reflect a small portion of the justification. (2) Domain adaptation and transfer learning enable AI models to be trained and applied across multiple domains. For example, a classification task based on images acquired on different acquisition hardware. (3) Federated learning enables learning large-scale models without exposing sensitive personal health information. Unlike centralized AI learning, where the centralized learning machine has access to the entire training data, the federated learning process iteratively updates models across multiple sites by exchanging only parameter updates, not personal health data. This narrative review covers the basic concepts, highlights relevant corner-stone and state-of-the-art research in the field, and discusses perspectives.
Applications of Generative Adversarial Networks in Neuroimaging and Clinical Neuroscience
Jun 14, 2022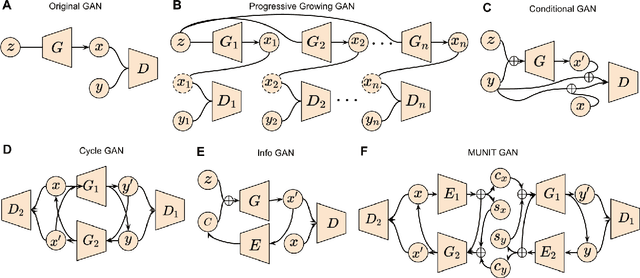
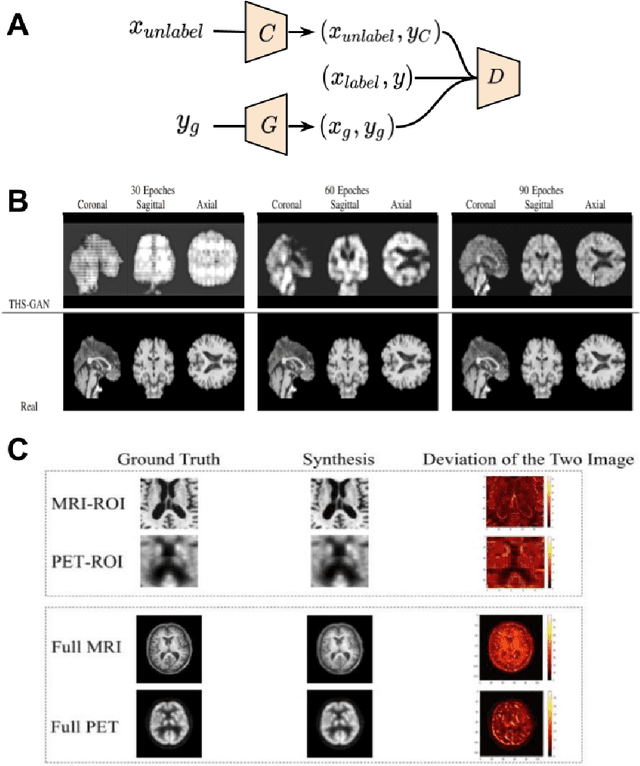
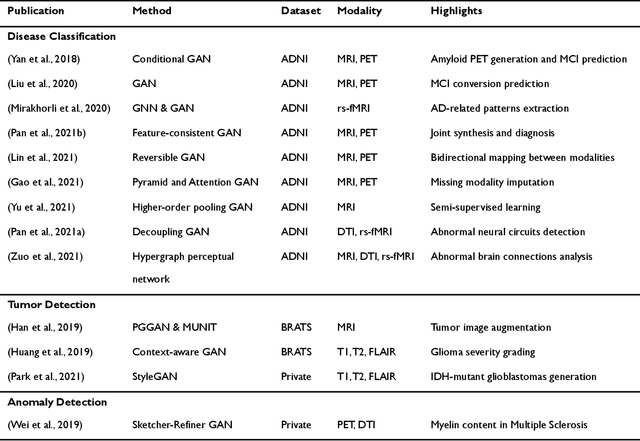
Abstract:Generative adversarial networks (GANs) are one powerful type of deep learning models that have been successfully utilized in numerous fields. They belong to a broader family called generative methods, which generate new data with a probabilistic model by learning sample distribution from real examples. In the clinical context, GANs have shown enhanced capabilities in capturing spatially complex, nonlinear, and potentially subtle disease effects compared to traditional generative methods. This review appraises the existing literature on the applications of GANs in imaging studies of various neurological conditions, including Alzheimer's disease, brain tumors, brain aging, and multiple sclerosis. We provide an intuitive explanation of various GAN methods for each application and further discuss the main challenges, open questions, and promising future directions of leveraging GANs in neuroimaging. We aim to bridge the gap between advanced deep learning methods and neurology research by highlighting how GANs can be leveraged to support clinical decision making and contribute to a better understanding of the structural and functional patterns of brain diseases.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021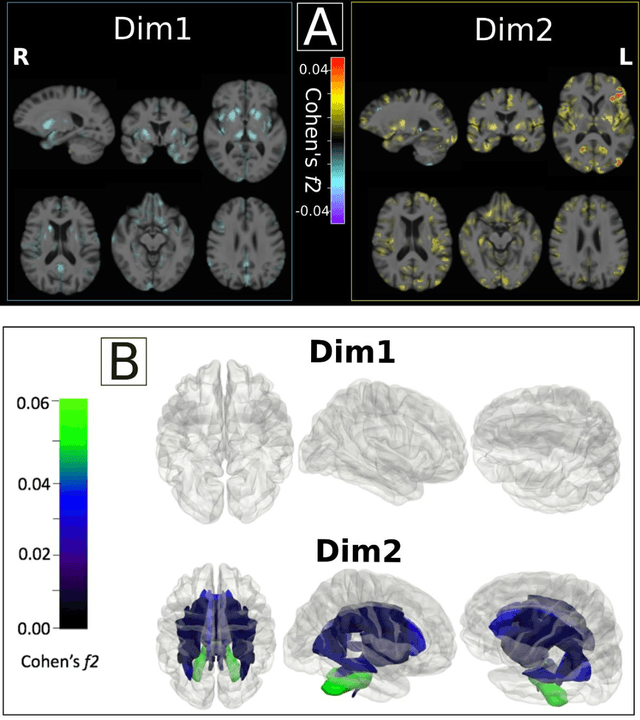
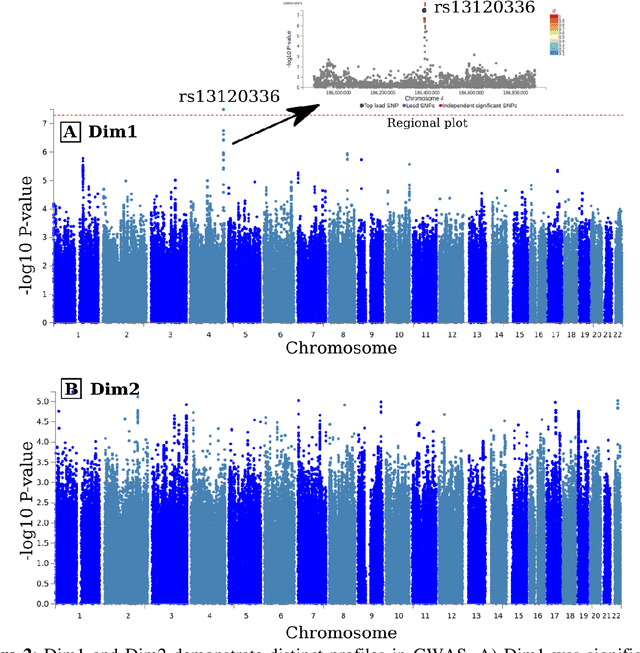
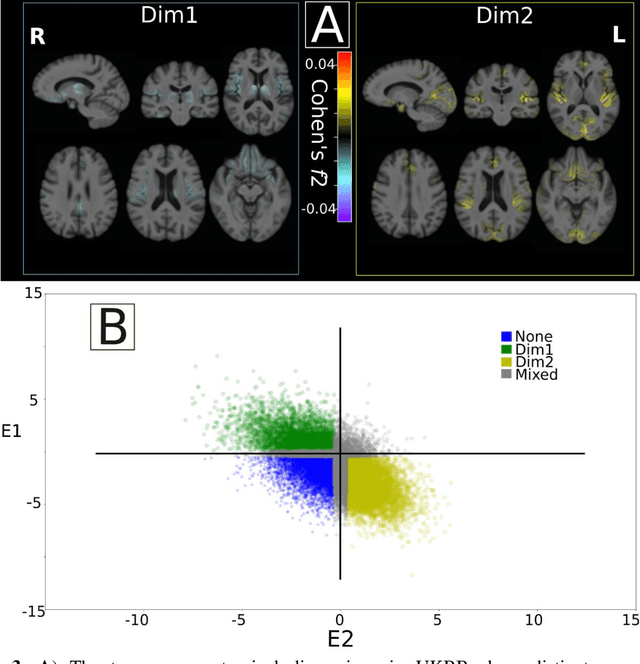
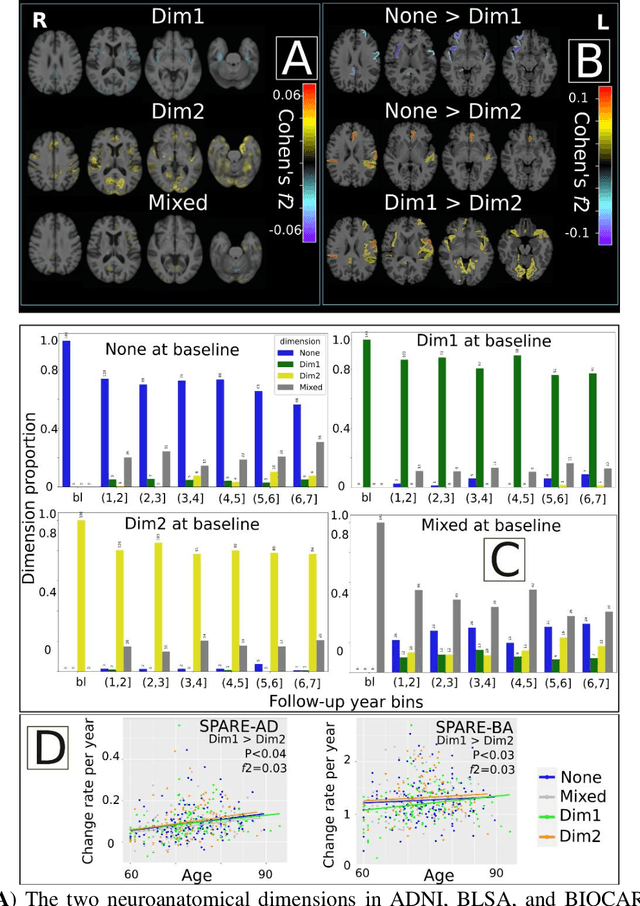
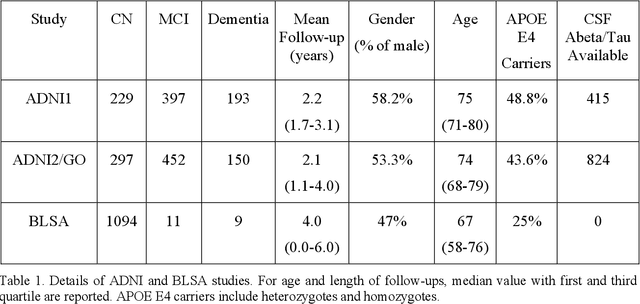
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
Disentangling Alzheimer's disease neurodegeneration from typical brain aging using machine learning
Sep 08, 2021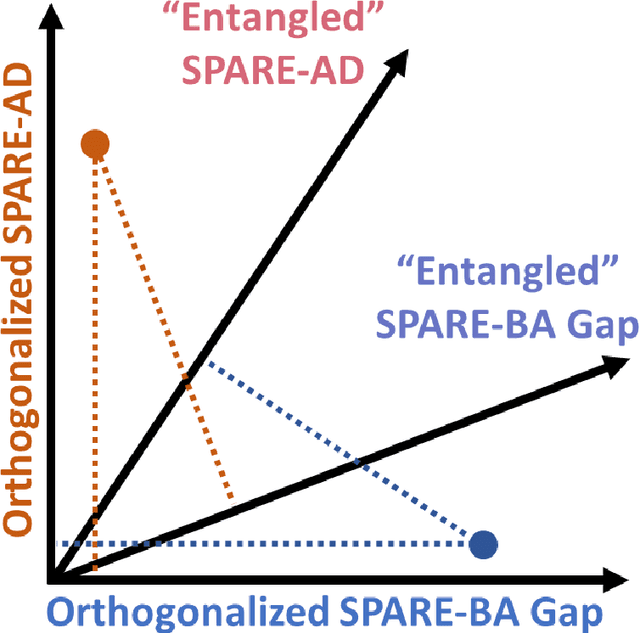
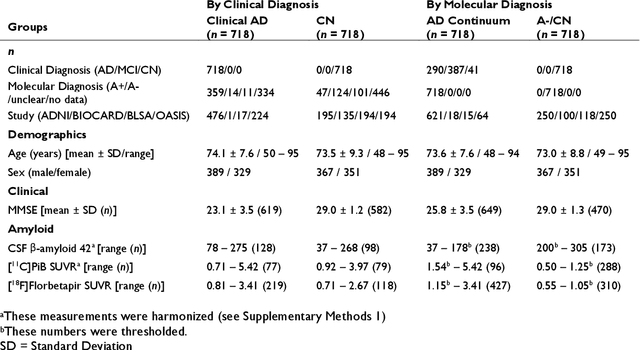
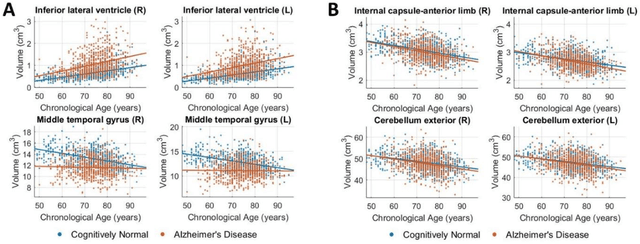
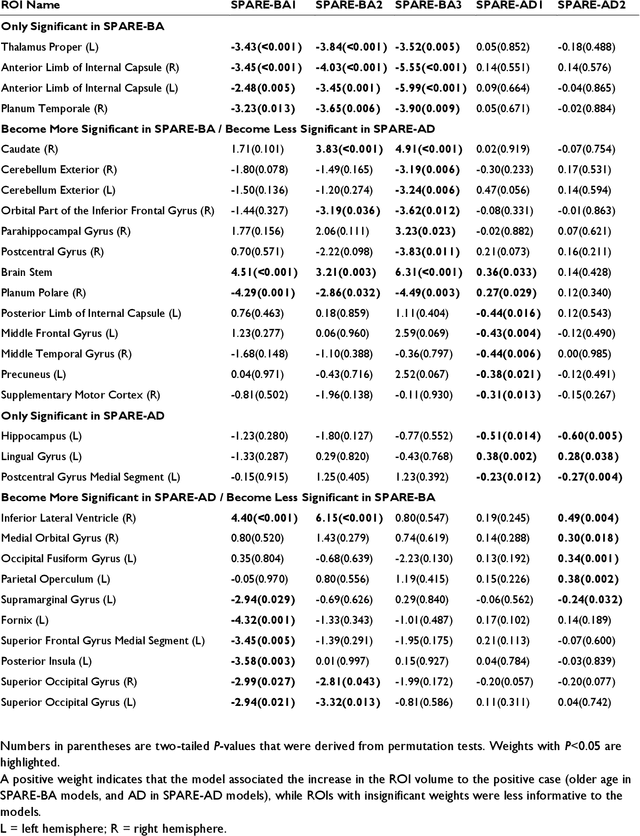
Abstract:Neuroimaging biomarkers that distinguish between typical brain aging and Alzheimer's disease (AD) are valuable for determining how much each contributes to cognitive decline. Machine learning models can derive multi-variate brain change patterns related to the two processes, including the SPARE-AD (Spatial Patterns of Atrophy for Recognition of Alzheimer's Disease) and SPARE-BA (of Brain Aging) investigated herein. However, substantial overlap between brain regions affected in the two processes confounds measuring them independently. We present a methodology toward disentangling the two. T1-weighted MRI images of 4,054 participants (48-95 years) with AD, mild cognitive impairment (MCI), or cognitively normal (CN) diagnoses from the iSTAGING (Imaging-based coordinate SysTem for AGIng and NeurodeGenerative diseases) consortium were analyzed. First, a subset of AD patients and CN adults were selected based purely on clinical diagnoses to train SPARE-BA1 (regression of age using CN individuals) and SPARE-AD1 (classification of CN versus AD). Second, analogous groups were selected based on clinical and molecular markers to train SPARE-BA2 and SPARE-AD2: amyloid-positive (A+) AD continuum group (consisting of A+AD, A+MCI, and A+ and tau-positive CN individuals) and amyloid-negative (A-) CN group. Finally, the combined group of the AD continuum and A-/CN individuals was used to train SPARE-BA3, with the intention to estimate brain age regardless of AD-related brain changes. Disentangled SPARE models derived brain patterns that were more specific to the two types of the brain changes. Correlation between the SPARE-BA and SPARE-AD was significantly reduced. Correlation of disentangled SPARE-AD was non-inferior to the molecular measurements and to the number of APOE4 alleles, but was less to AD-related psychometric test scores, suggesting contribution of advanced brain aging to these scores.
Disentangling brain heterogeneity via semi-supervised deep-learning and MRI: dimensional representations of Alzheimer's Disease
Feb 24, 2021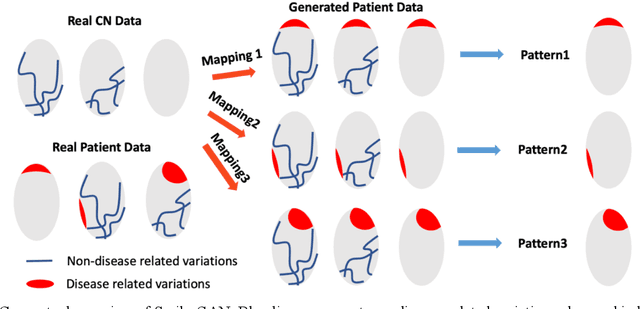
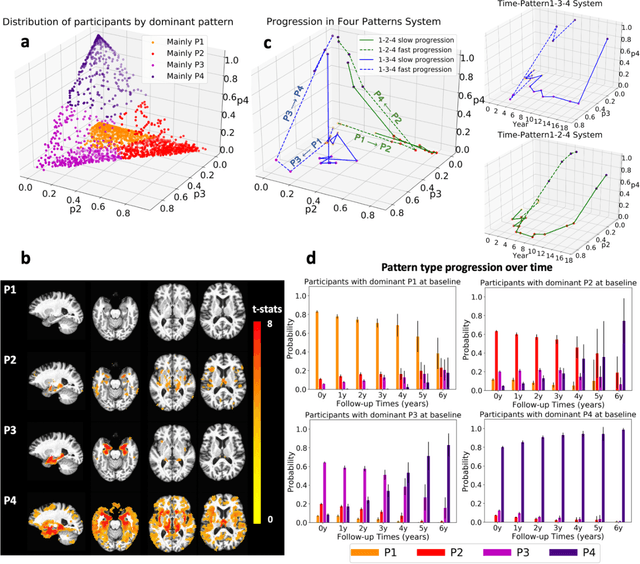
Abstract:Heterogeneity of brain diseases is a challenge for precision diagnosis/prognosis. We describe and validate Smile-GAN (SeMI-supervised cLustEring-Generative Adversarial Network), a novel semi-supervised deep-clustering method, which dissects neuroanatomical heterogeneity, enabling identification of disease subtypes via their imaging signatures relative to controls. When applied to MRIs (2 studies; 2,832 participants; 8,146 scans) including cognitively normal individuals and those with cognitive impairment and dementia, Smile-GAN identified 4 neurodegenerative patterns/axes: P1, normal anatomy and highest cognitive performance; P2, mild/diffuse atrophy and more prominent executive dysfunction; P3, focal medial temporal atrophy and relatively greater memory impairment; P4, advanced neurodegeneration. Further application to longitudinal data revealed two distinct progression pathways: P1$\rightarrow$P2$\rightarrow$P4 and P1$\rightarrow$P3$\rightarrow$P4. Baseline expression of these patterns predicted the pathway and rate of future neurodegeneration. Pattern expression offered better yet complementary performance in predicting clinical progression, compared to amyloid/tau. These deep-learning derived biomarkers offer promise for precision diagnostics and targeted clinical trial recruitment.
Medical Image Harmonization Using Deep Learning Based Canonical Mapping: Toward Robust and Generalizable Learning in Imaging
Oct 11, 2020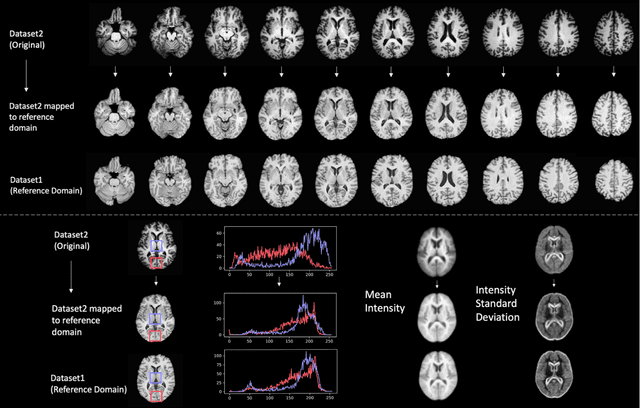
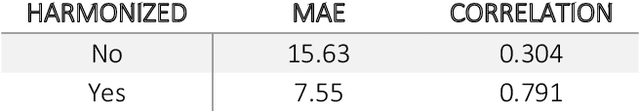
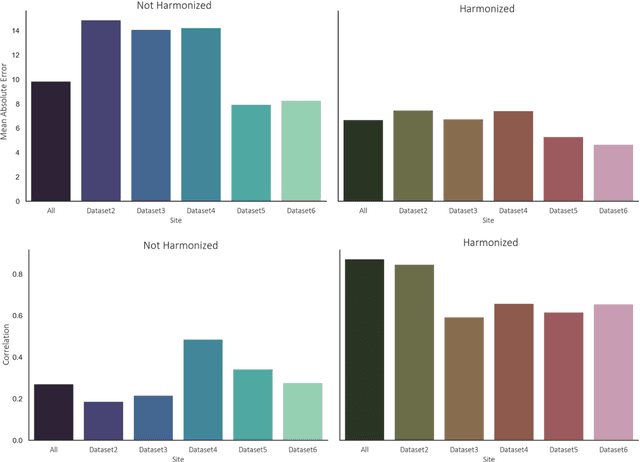
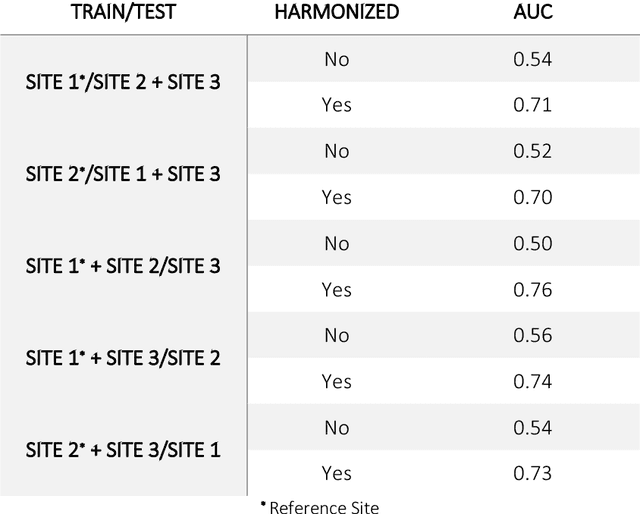
Abstract:Conventional and deep learning-based methods have shown great potential in the medical imaging domain, as means for deriving diagnostic, prognostic, and predictive biomarkers, and by contributing to precision medicine. However, these methods have yet to see widespread clinical adoption, in part due to limited generalization performance across various imaging devices, acquisition protocols, and patient populations. In this work, we propose a new paradigm in which data from a diverse range of acquisition conditions are "harmonized" to a common reference domain, where accurate model learning and prediction can take place. By learning an unsupervised image to image canonical mapping from diverse datasets to a reference domain using generative deep learning models, we aim to reduce confounding data variation while preserving semantic information, thereby rendering the learning task easier in the reference domain. We test this approach on two example problems, namely MRI-based brain age prediction and classification of schizophrenia, leveraging pooled cohorts of neuroimaging MRI data spanning 9 sites and 9701 subjects. Our results indicate a substantial improvement in these tasks in out-of-sample data, even when training is restricted to a single site.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge