Dhivya Srinivasan
from the iSTAGING consortium, for the ADNI
Gene-SGAN: a method for discovering disease subtypes with imaging and genetic signatures via multi-view weakly-supervised deep clustering
Jan 25, 2023


Abstract:Disease heterogeneity has been a critical challenge for precision diagnosis and treatment, especially in neurologic and neuropsychiatric diseases. Many diseases can display multiple distinct brain phenotypes across individuals, potentially reflecting disease subtypes that can be captured using MRI and machine learning methods. However, biological interpretability and treatment relevance are limited if the derived subtypes are not associated with genetic drivers or susceptibility factors. Herein, we describe Gene-SGAN - a multi-view, weakly-supervised deep clustering method - which dissects disease heterogeneity by jointly considering phenotypic and genetic data, thereby conferring genetic correlations to the disease subtypes and associated endophenotypic signatures. We first validate the generalizability, interpretability, and robustness of Gene-SGAN in semi-synthetic experiments. We then demonstrate its application to real multi-site datasets from 28,858 individuals, deriving subtypes of Alzheimer's disease and brain endophenotypes associated with hypertension, from MRI and SNP data. Derived brain phenotypes displayed significant differences in neuroanatomical patterns, genetic determinants, biological and clinical biomarkers, indicating potentially distinct underlying neuropathologic processes, genetic drivers, and susceptibility factors. Overall, Gene-SGAN is broadly applicable to disease subtyping and endophenotype discovery, and is herein tested on disease-related, genetically-driven neuroimaging phenotypes.
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021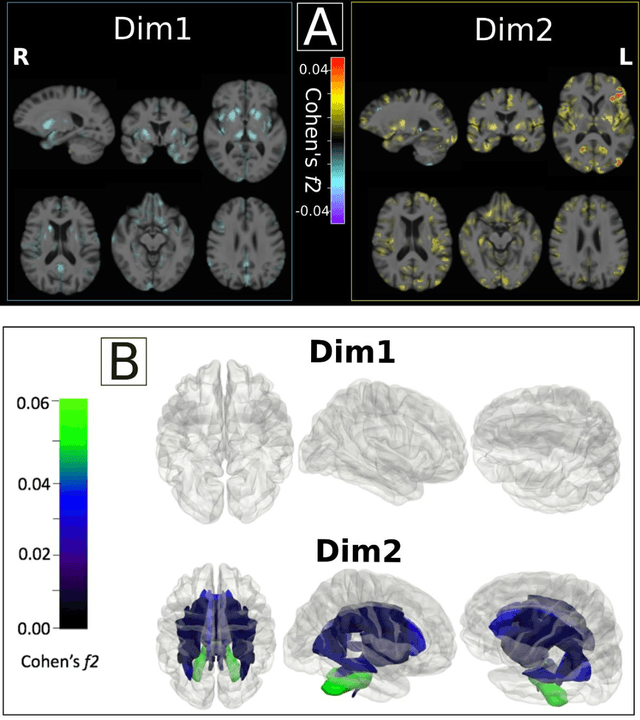
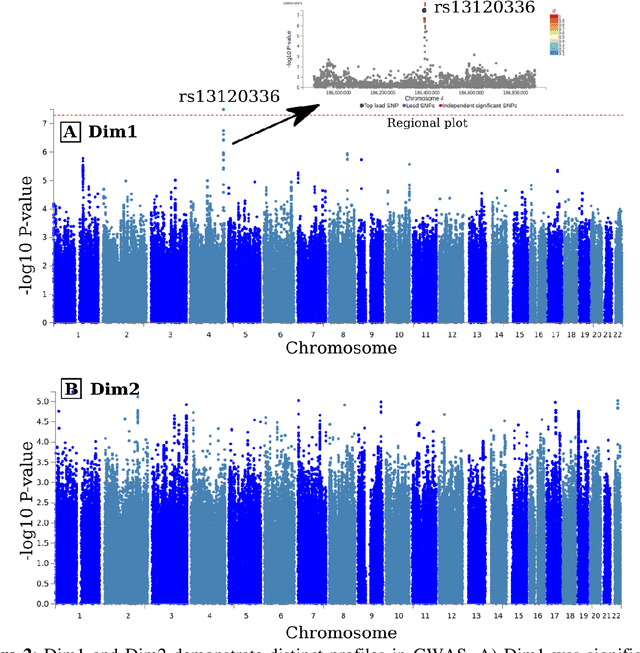
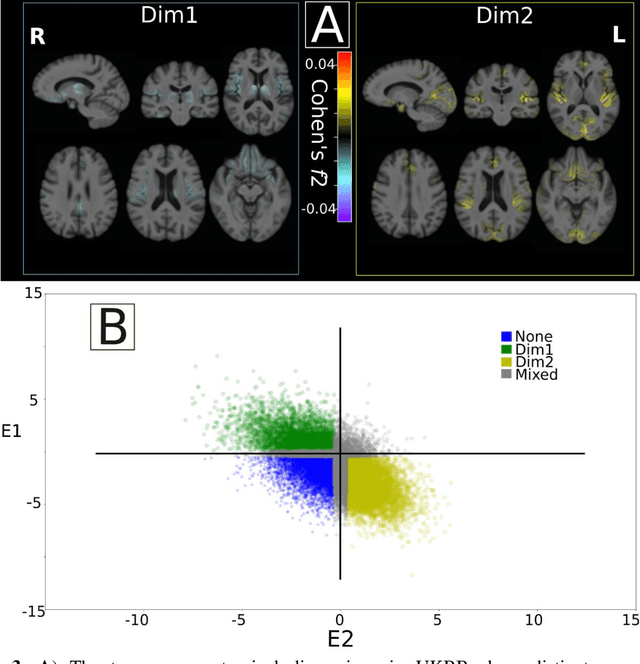
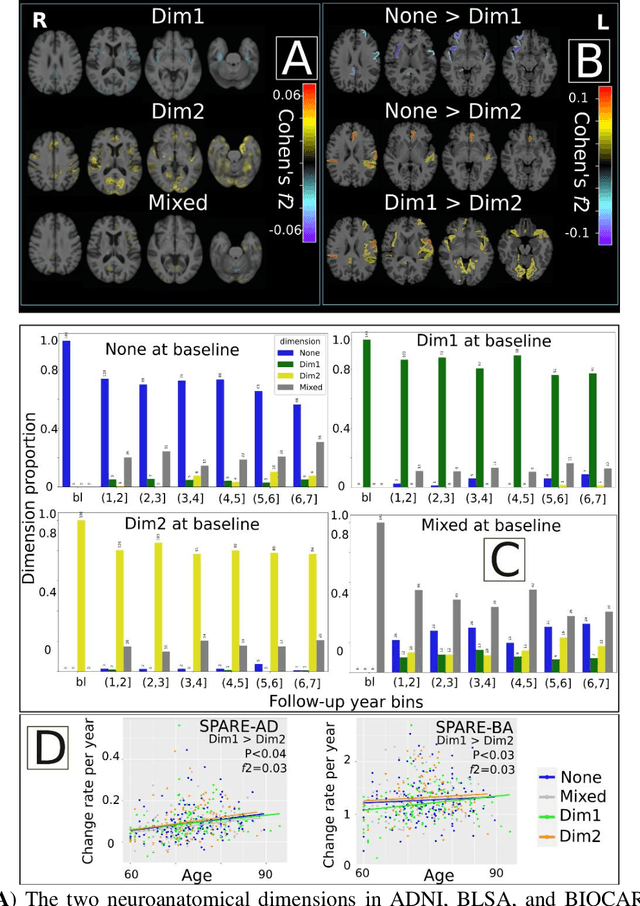
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
Disentangling Alzheimer's disease neurodegeneration from typical brain aging using machine learning
Sep 08, 2021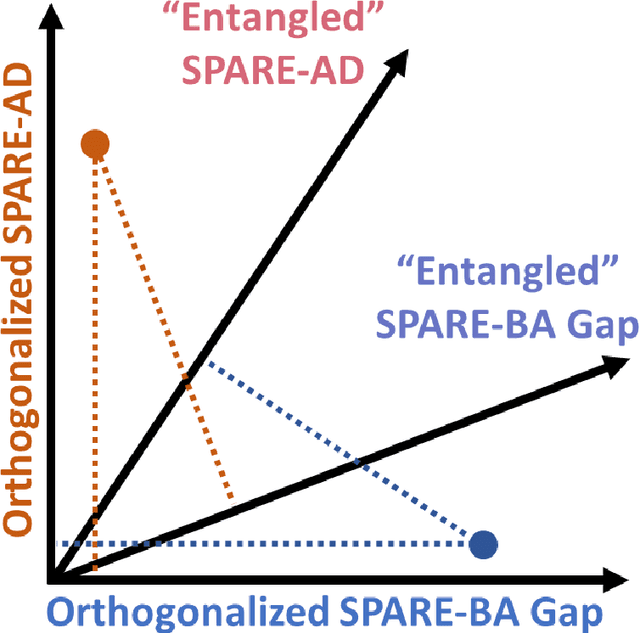
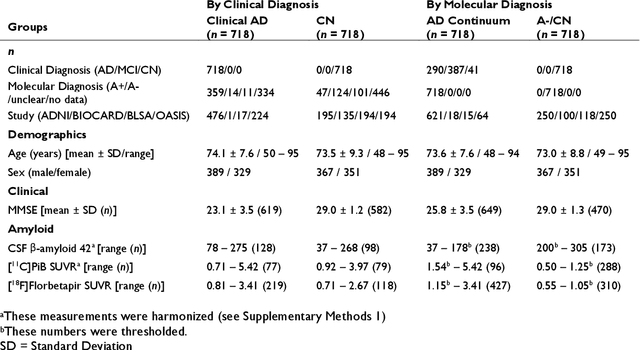
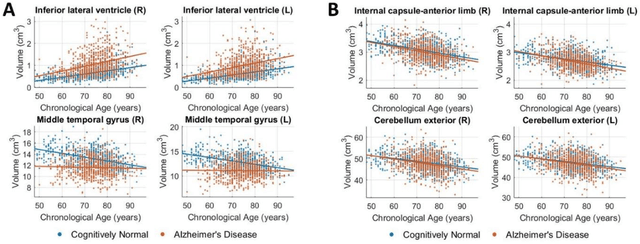
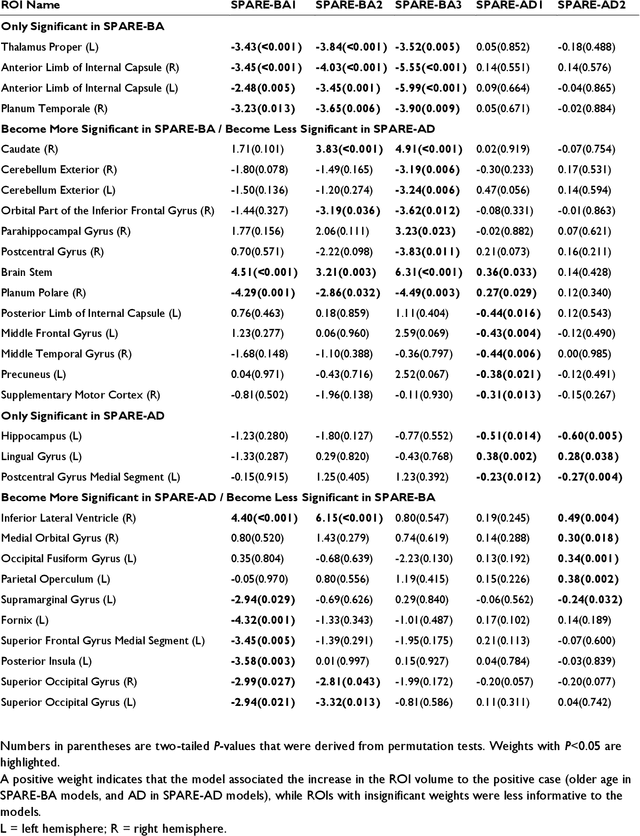
Abstract:Neuroimaging biomarkers that distinguish between typical brain aging and Alzheimer's disease (AD) are valuable for determining how much each contributes to cognitive decline. Machine learning models can derive multi-variate brain change patterns related to the two processes, including the SPARE-AD (Spatial Patterns of Atrophy for Recognition of Alzheimer's Disease) and SPARE-BA (of Brain Aging) investigated herein. However, substantial overlap between brain regions affected in the two processes confounds measuring them independently. We present a methodology toward disentangling the two. T1-weighted MRI images of 4,054 participants (48-95 years) with AD, mild cognitive impairment (MCI), or cognitively normal (CN) diagnoses from the iSTAGING (Imaging-based coordinate SysTem for AGIng and NeurodeGenerative diseases) consortium were analyzed. First, a subset of AD patients and CN adults were selected based purely on clinical diagnoses to train SPARE-BA1 (regression of age using CN individuals) and SPARE-AD1 (classification of CN versus AD). Second, analogous groups were selected based on clinical and molecular markers to train SPARE-BA2 and SPARE-AD2: amyloid-positive (A+) AD continuum group (consisting of A+AD, A+MCI, and A+ and tau-positive CN individuals) and amyloid-negative (A-) CN group. Finally, the combined group of the AD continuum and A-/CN individuals was used to train SPARE-BA3, with the intention to estimate brain age regardless of AD-related brain changes. Disentangled SPARE models derived brain patterns that were more specific to the two types of the brain changes. Correlation between the SPARE-BA and SPARE-AD was significantly reduced. Correlation of disentangled SPARE-AD was non-inferior to the molecular measurements and to the number of APOE4 alleles, but was less to AD-related psychometric test scores, suggesting contribution of advanced brain aging to these scores.
Medical Image Harmonization Using Deep Learning Based Canonical Mapping: Toward Robust and Generalizable Learning in Imaging
Oct 11, 2020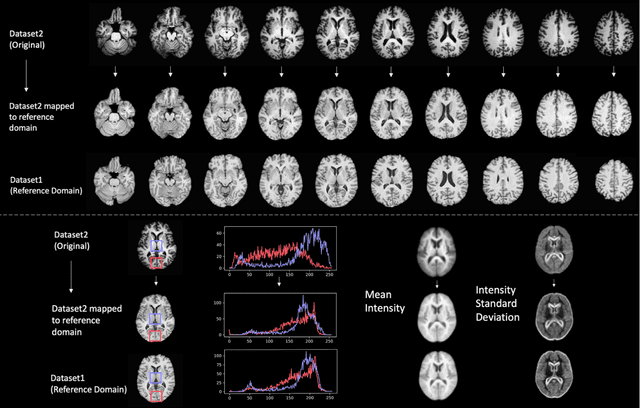
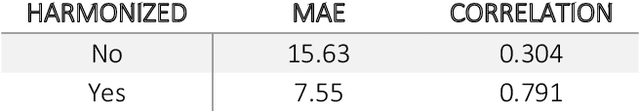
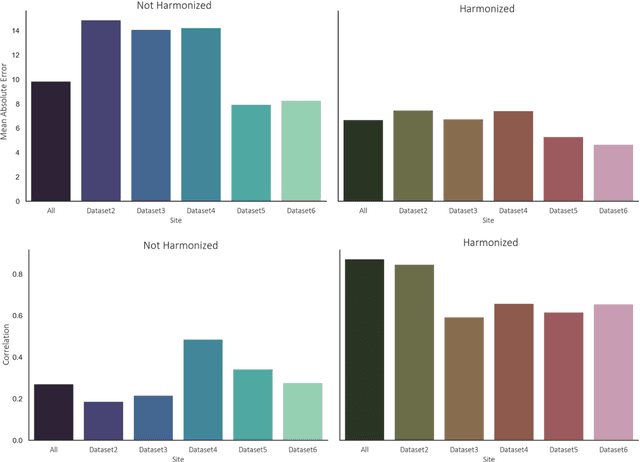
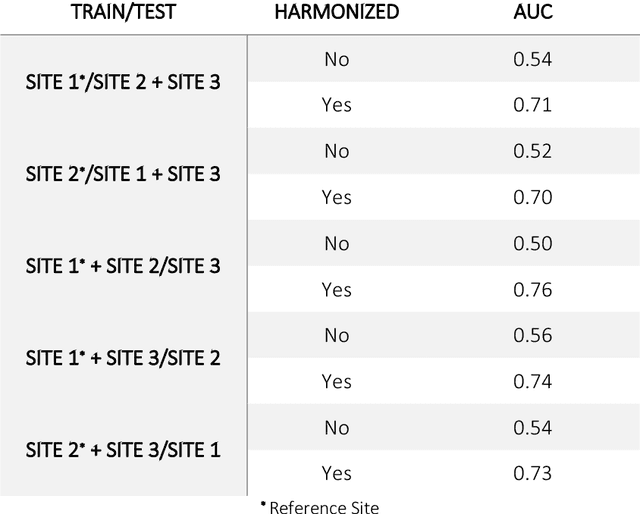
Abstract:Conventional and deep learning-based methods have shown great potential in the medical imaging domain, as means for deriving diagnostic, prognostic, and predictive biomarkers, and by contributing to precision medicine. However, these methods have yet to see widespread clinical adoption, in part due to limited generalization performance across various imaging devices, acquisition protocols, and patient populations. In this work, we propose a new paradigm in which data from a diverse range of acquisition conditions are "harmonized" to a common reference domain, where accurate model learning and prediction can take place. By learning an unsupervised image to image canonical mapping from diverse datasets to a reference domain using generative deep learning models, we aim to reduce confounding data variation while preserving semantic information, thereby rendering the learning task easier in the reference domain. We test this approach on two example problems, namely MRI-based brain age prediction and classification of schizophrenia, leveraging pooled cohorts of neuroimaging MRI data spanning 9 sites and 9701 subjects. Our results indicate a substantial improvement in these tasks in out-of-sample data, even when training is restricted to a single site.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge