cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Beyond Occlusion: In Search for Near Real-Time Explainability of CNN-Based Prostate Cancer Classification
Dec 19, 2025



Deep neural networks are starting to show their worth in critical applications such as assisted cancer diagnosis. However, for their outputs to get accepted in practice, the results they provide should be explainable in a way easily understood by pathologists. A well-known and widely used explanation technique is occlusion, which, however, can take a long time to compute, thus slowing the development and interaction with pathologists. In this work, we set out to find a faster replacement for occlusion in a successful system for detecting prostate cancer. Since there is no established framework for comparing the performance of various explanation methods, we first identified suitable comparison criteria and selected corresponding metrics. Based on the results, we were able to choose a different explanation method, which cut the previously required explanation time at least by a factor of 10, without any negative impact on the quality of outputs. This speedup enables rapid iteration in model development and debugging and brings us closer to adopting AI-assisted prostate cancer detection in clinical settings. We propose that our approach to finding the replacement for occlusion can be used to evaluate candidate methods in other related applications.
Agent-Based Output Drift Detection for Breast Cancer Response Prediction in a Multisite Clinical Decision Support System
Dec 20, 2025Modern clinical decision support systems can concurrently serve multiple, independent medical imaging institutions, but their predictive performance may degrade across sites due to variations in patient populations, imaging hardware, and acquisition protocols. Continuous surveillance of predictive model outputs offers a safe and reliable approach for identifying such distributional shifts without ground truth labels. However, most existing methods rely on centralized monitoring of aggregated predictions, overlooking site-specific drift dynamics. We propose an agent-based framework for detecting drift and assessing its severity in multisite clinical AI systems. To evaluate its effectiveness, we simulate a multi-center environment for output-based drift detection, assigning each site a drift monitoring agent that performs batch-wise comparisons of model outputs against a reference distribution. We analyse several multi-center monitoring schemes, that differ in how the reference is obtained (site-specific, global, production-only and adaptive), alongside a centralized baseline. Results on real-world breast cancer imaging data using a pathological complete response prediction model shows that all multi-center schemes outperform centralized monitoring, with F1-score improvements up to 10.3% in drift detection. In the absence of site-specific references, the adaptive scheme performs best, with F1-scores of 74.3% for drift detection and 83.7% for drift severity classification. These findings suggest that adaptive, site-aware agent-based drift monitoring can enhance reliability of multisite clinical decision support systems.
Fairness Evaluation of Risk Estimation Models for Lung Cancer Screening
Dec 23, 2025Lung cancer is the leading cause of cancer-related mortality in adults worldwide. Screening high-risk individuals with annual low-dose CT (LDCT) can support earlier detection and reduce deaths, but widespread implementation may strain the already limited radiology workforce. AI models have shown potential in estimating lung cancer risk from LDCT scans. However, high-risk populations for lung cancer are diverse, and these models' performance across demographic groups remains an open question. In this study, we drew on the considerations on confounding factors and ethically significant biases outlined in the JustEFAB framework to evaluate potential performance disparities and fairness in two deep learning risk estimation models for lung cancer screening: the Sybil lung cancer risk model and the Venkadesh21 nodule risk estimator. We also examined disparities in the PanCan2b logistic regression model recommended in the British Thoracic Society nodule management guideline. Both deep learning models were trained on data from the US-based National Lung Screening Trial (NLST), and assessed on a held-out NLST validation set. We evaluated AUROC, sensitivity, and specificity across demographic subgroups, and explored potential confounding from clinical risk factors. We observed a statistically significant AUROC difference in Sybil's performance between women (0.88, 95% CI: 0.86, 0.90) and men (0.81, 95% CI: 0.78, 0.84, p < .001). At 90% specificity, Venkadesh21 showed lower sensitivity for Black (0.39, 95% CI: 0.23, 0.59) than White participants (0.69, 95% CI: 0.65, 0.73). These differences were not explained by available clinical confounders and thus may be classified as unfair biases according to JustEFAB. Our findings highlight the importance of improving and monitoring model performance across underrepresented subgroups, and further research on algorithmic fairness, in lung cancer screening.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:025
DBT-DINO: Towards Foundation model based analysis of Digital Breast Tomosynthesis
Dec 15, 2025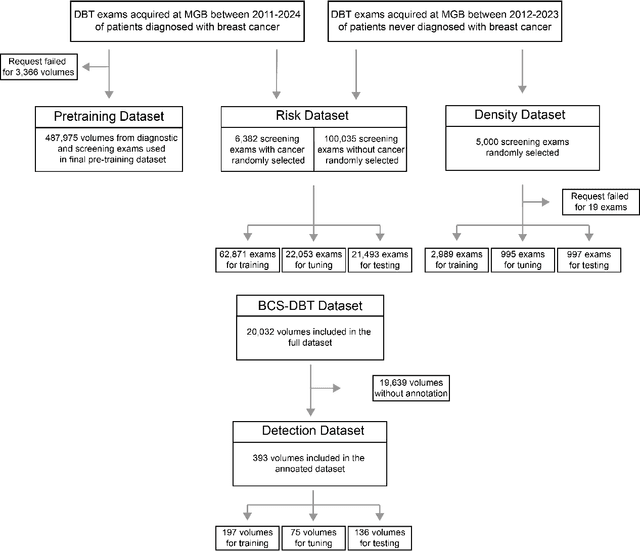
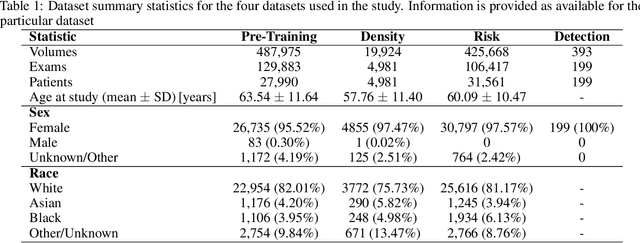
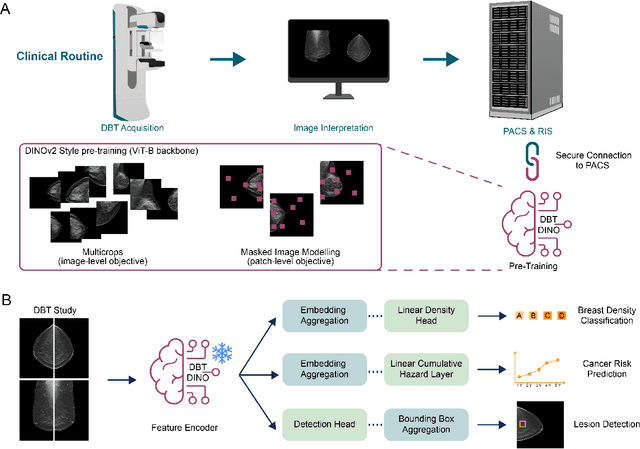
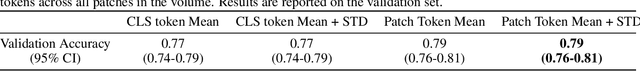
Foundation models have shown promise in medical imaging but remain underexplored for three-dimensional imaging modalities. No foundation model currently exists for Digital Breast Tomosynthesis (DBT), despite its use for breast cancer screening. To develop and evaluate a foundation model for DBT (DBT-DINO) across multiple clinical tasks and assess the impact of domain-specific pre-training. Self-supervised pre-training was performed using the DINOv2 methodology on over 25 million 2D slices from 487,975 DBT volumes from 27,990 patients. Three downstream tasks were evaluated: (1) breast density classification using 5,000 screening exams; (2) 5-year risk of developing breast cancer using 106,417 screening exams; and (3) lesion detection using 393 annotated volumes. For breast density classification, DBT-DINO achieved an accuracy of 0.79 (95\% CI: 0.76--0.81), outperforming both the MetaAI DINOv2 baseline (0.73, 95\% CI: 0.70--0.76, p<.001) and DenseNet-121 (0.74, 95\% CI: 0.71--0.76, p<.001). For 5-year breast cancer risk prediction, DBT-DINO achieved an AUROC of 0.78 (95\% CI: 0.76--0.80) compared to DINOv2's 0.76 (95\% CI: 0.74--0.78, p=.57). For lesion detection, DINOv2 achieved a higher average sensitivity of 0.67 (95\% CI: 0.60--0.74) compared to DBT-DINO with 0.62 (95\% CI: 0.53--0.71, p=.60). DBT-DINO demonstrated better performance on cancerous lesions specifically with a detection rate of 78.8\% compared to Dinov2's 77.3\%. Using a dataset of unprecedented size, we developed DBT-DINO, the first foundation model for DBT. DBT-DINO demonstrated strong performance on breast density classification and cancer risk prediction. However, domain-specific pre-training showed variable benefits on the detection task, with ImageNet baseline outperforming DBT-DINO on general lesion detection, indicating that localized detection tasks require further methodological development.
LDP: Parameter-Efficient Fine-Tuning of Multimodal LLM for Medical Report Generation
Dec 11, 2025Colonoscopic polyp diagnosis is pivotal for early colorectal cancer detection, yet traditional automated reporting suffers from inconsistencies and hallucinations due to the scarcity of high-quality multimodal medical data. To bridge this gap, we propose LDP, a novel framework leveraging multimodal large language models (MLLMs) for professional polyp diagnosis report generation. Specifically, we curate MMEndo, a multimodal endoscopic dataset comprising expert-annotated colonoscopy image-text pairs. We fine-tune the Qwen2-VL-7B backbone using Parameter-Efficient Fine-Tuning (LoRA) and align it with clinical standards via Direct Preference Optimization (DPO). Extensive experiments show that our LDP outperforms existing baselines on both automated metrics and rigorous clinical expert evaluations (achieving a Physician Score of 7.2/10), significantly reducing training computational costs by 833x compared to full fine-tuning. The proposed solution offers a scalable, clinically viable path for primary healthcare, with additional validation on the IU-XRay dataset confirming its robustness.
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Tumor-anchored deep feature random forests for out-of-distribution detection in lung cancer segmentation
Dec 09, 2025Accurate segmentation of cancerous lesions from 3D computed tomography (CT) scans is essential for automated treatment planning and response assessment. However, even state-of-the-art models combining self-supervised learning (SSL) pretrained transformers with convolutional decoders are susceptible to out-of-distribution (OOD) inputs, generating confidently incorrect tumor segmentations, posing risks for safe clinical deployment. Existing logit-based methods suffer from task-specific model biases, while architectural enhancements to explicitly detect OOD increase parameters and computational costs. Hence, we introduce a plug-and-play and lightweight post-hoc random forests-based OOD detection framework called RF-Deep that leverages deep features with limited outlier exposure. RF-Deep enhances generalization to imaging variations by repurposing the hierarchical features from the pretrained-then-finetuned backbone encoder, providing task-relevant OOD detection by extracting the features from multiple regions of interest anchored to the predicted tumor segmentations. Hence, it scales to images of varying fields-of-view. We compared RF-Deep against existing OOD detection methods using 1,916 CT scans across near-OOD (pulmonary embolism, negative COVID-19) and far-OOD (kidney cancer, healthy pancreas) datasets. RF-Deep achieved AUROC > 93.50 for the challenging near-OOD datasets and near-perfect detection (AUROC > 99.00) for the far-OOD datasets, substantially outperforming logit-based and radiomics approaches. RF-Deep maintained similar performance consistency across networks of different depths and pretraining strategies, demonstrating its effectiveness as a lightweight, architecture-agnostic approach to enhance the reliability of tumor segmentation from CT volumes.
From SAM to DINOv2: Towards Distilling Foundation Models to Lightweight Baselines for Generalized Polyp Segmentation
Dec 10, 2025Accurate polyp segmentation during colonoscopy is critical for the early detection of colorectal cancer and still remains challenging due to significant size, shape, and color variations, and the camouflaged nature of polyps. While lightweight baseline models such as U-Net, U-Net++, and PraNet offer advantages in terms of easy deployment and low computational cost, they struggle to deal with the above issues, leading to limited segmentation performance. In contrast, large-scale vision foundation models such as SAM, DINOv2, OneFormer, and Mask2Former have exhibited impressive generalization performance across natural image domains. However, their direct transfer to medical imaging tasks (e.g., colonoscopic polyp segmentation) is not straightforward, primarily due to the scarcity of large-scale datasets and lack of domain-specific knowledge. To bridge this gap, we propose a novel distillation framework, Polyp-DiFoM, that transfers the rich representations of foundation models into lightweight segmentation baselines, allowing efficient and accurate deployment in clinical settings. In particular, we infuse semantic priors from the foundation models into canonical architectures such as U-Net and U-Net++ and further perform frequency domain encoding for enhanced distillation, corroborating their generalization capability. Extensive experiments are performed across five benchmark datasets, such as Kvasir-SEG, CVC-ClinicDB, ETIS, ColonDB, and CVC-300. Notably, Polyp-DiFoM consistently outperforms respective baseline models significantly, as well as the state-of-the-art model, with nearly 9 times reduced computation overhead. The code is available at https://github.com/lostinrepo/PolypDiFoM.
General OOD Detection via Model-aware and Subspace-aware Variable Priority
Dec 15, 2025Out-of-distribution (OOD) detection is essential for determining when a supervised model encounters inputs that differ meaningfully from its training distribution. While widely studied in classification, OOD detection for regression and survival analysis remains limited due to the absence of discrete labels and the challenge of quantifying predictive uncertainty. We introduce a framework for OOD detection that is simultaneously model aware and subspace aware, and that embeds variable prioritization directly into the detection step. The method uses the fitted predictor to construct localized neighborhoods around each test case that emphasize the features driving the model's learned relationship and downweight directions that are less relevant to prediction. It produces OOD scores without relying on global distance metrics or estimating the full feature density. The framework is applicable across outcome types, and in our implementation we use random forests, where the rule structure yields transparent neighborhoods and effective scoring. Experiments on synthetic and real data benchmarks designed to isolate functional shifts show consistent improvements over existing methods. We further demonstrate the approach in an esophageal cancer survival study, where distribution shifts related to lymphadenectomy identify patterns relevant to surgical guidelines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge