Aneesh Rangnekar
MHub.ai: A Simple, Standardized, and Reproducible Platform for AI Models in Medical Imaging
Jan 15, 2026Abstract:Artificial intelligence (AI) has the potential to transform medical imaging by automating image analysis and accelerating clinical research. However, research and clinical use are limited by the wide variety of AI implementations and architectures, inconsistent documentation, and reproducibility issues. Here, we introduce MHub.ai, an open-source, container-based platform that standardizes access to AI models with minimal configuration, promoting accessibility and reproducibility in medical imaging. MHub.ai packages models from peer-reviewed publications into standardized containers that support direct processing of DICOM and other formats, provide a unified application interface, and embed structured metadata. Each model is accompanied by publicly available reference data that can be used to confirm model operation. MHub.ai includes an initial set of state-of-the-art segmentation, prediction, and feature extraction models for different modalities. The modular framework enables adaptation of any model and supports community contributions. We demonstrate the utility of the platform in a clinical use case through comparative evaluation of lung segmentation models. To further strengthen transparency and reproducibility, we publicly release the generated segmentations and evaluation metrics and provide interactive dashboards that allow readers to inspect individual cases and reproduce or extend our analysis. By simplifying model use, MHub.ai enables side-by-side benchmarking with identical execution commands and standardized outputs, and lowers the barrier to clinical translation.
brat: Aligned Multi-View Embeddings for Brain MRI Analysis
Dec 21, 2025



Abstract:We present brat (brain report alignment transformer), a multi-view representation learning framework for brain magnetic resonance imaging (MRI) trained on MRIs paired with clinical reports. Brain MRIs present unique challenges due to the presence of numerous, highly varied, and often subtle abnormalities that are localized to a few slices within a 3D volume. To address these challenges, we introduce a brain MRI dataset $10\times$ larger than existing ones, containing approximately 80,000 3D scans with corresponding radiology reports, and propose a multi-view pre-training approach inspired by advances in document retrieval. We develop an implicit query-feature matching mechanism and adopt concepts from quality-diversity to obtain multi-view embeddings of MRIs that are aligned with the clinical features given by report sentences. We evaluate our approach across multiple vision-language and vision tasks, demonstrating substantial performance improvements. The brat foundation models are publicly released.
Tumor-anchored deep feature random forests for out-of-distribution detection in lung cancer segmentation
Dec 09, 2025Abstract:Accurate segmentation of cancerous lesions from 3D computed tomography (CT) scans is essential for automated treatment planning and response assessment. However, even state-of-the-art models combining self-supervised learning (SSL) pretrained transformers with convolutional decoders are susceptible to out-of-distribution (OOD) inputs, generating confidently incorrect tumor segmentations, posing risks for safe clinical deployment. Existing logit-based methods suffer from task-specific model biases, while architectural enhancements to explicitly detect OOD increase parameters and computational costs. Hence, we introduce a plug-and-play and lightweight post-hoc random forests-based OOD detection framework called RF-Deep that leverages deep features with limited outlier exposure. RF-Deep enhances generalization to imaging variations by repurposing the hierarchical features from the pretrained-then-finetuned backbone encoder, providing task-relevant OOD detection by extracting the features from multiple regions of interest anchored to the predicted tumor segmentations. Hence, it scales to images of varying fields-of-view. We compared RF-Deep against existing OOD detection methods using 1,916 CT scans across near-OOD (pulmonary embolism, negative COVID-19) and far-OOD (kidney cancer, healthy pancreas) datasets. RF-Deep achieved AUROC > 93.50 for the challenging near-OOD datasets and near-perfect detection (AUROC > 99.00) for the far-OOD datasets, substantially outperforming logit-based and radiomics approaches. RF-Deep maintained similar performance consistency across networks of different depths and pretraining strategies, demonstrating its effectiveness as a lightweight, architecture-agnostic approach to enhance the reliability of tumor segmentation from CT volumes.
Random forest-based out-of-distribution detection for robust lung cancer segmentation
Aug 26, 2025Abstract:Accurate detection and segmentation of cancerous lesions from computed tomography (CT) scans is essential for automated treatment planning and cancer treatment response assessment. Transformer-based models with self-supervised pretraining can produce reliably accurate segmentation from in-distribution (ID) data but degrade when applied to out-of-distribution (OOD) datasets. We address this challenge with RF-Deep, a random forest classifier that utilizes deep features from a pretrained transformer encoder of the segmentation model to detect OOD scans and enhance segmentation reliability. The segmentation model comprises a Swin Transformer encoder, pretrained with masked image modeling (SimMIM) on 10,432 unlabeled 3D CT scans covering cancerous and non-cancerous conditions, with a convolution decoder, trained to segment lung cancers in 317 3D scans. Independent testing was performed on 603 3D CT public datasets that included one ID dataset and four OOD datasets comprising chest CTs with pulmonary embolism (PE) and COVID-19, and abdominal CTs with kidney cancers and healthy volunteers. RF-Deep detected OOD cases with a FPR95 of 18.26%, 27.66%, and less than 0.1% on PE, COVID-19, and abdominal CTs, consistently outperforming established OOD approaches. The RF-Deep classifier provides a simple and effective approach to enhance reliability of cancer segmentation in ID and OOD scenarios.
Pretrained hybrid transformer for generalizable cardiac substructures segmentation from contrast and non-contrast CTs in lung and breast cancers
May 16, 2025Abstract:AI automated segmentations for radiation treatment planning (RTP) can deteriorate when applied in clinical cases with different characteristics than training dataset. Hence, we refined a pretrained transformer into a hybrid transformer convolutional network (HTN) to segment cardiac substructures lung and breast cancer patients acquired with varying imaging contrasts and patient scan positions. Cohort I, consisting of 56 contrast-enhanced (CECT) and 124 non-contrast CT (NCCT) scans from patients with non-small cell lung cancers acquired in supine position, was used to create oracle with all 180 training cases and balanced (CECT: 32, NCCT: 32 training) HTN models. Models were evaluated on a held-out validation set of 60 cohort I patients and 66 patients with breast cancer from cohort II acquired in supine (n=45) and prone (n=21) positions. Accuracy was measured using DSC, HD95, and dose metrics. Publicly available TotalSegmentator served as the benchmark. The oracle and balanced models were similarly accurate (DSC Cohort I: 0.80 \pm 0.10 versus 0.81 \pm 0.10; Cohort II: 0.77 \pm 0.13 versus 0.80 \pm 0.12), outperforming TotalSegmentator. The balanced model, using half the training cases as oracle, produced similar dose metrics as manual delineations for all cardiac substructures. This model was robust to CT contrast in 6 out of 8 substructures and patient scan position variations in 5 out of 8 substructures and showed low correlations of accuracy to patient size and age. A HTN demonstrated robustly accurate (geometric and dose metrics) cardiac substructures segmentation from CTs with varying imaging and patient characteristics, one key requirement for clinical use. Moreover, the model combining pretraining with balanced distribution of NCCT and CECT scans was able to provide reliably accurate segmentations under varied conditions with far fewer labeled datasets compared to an oracle model.
Transformer-based segmentation of adnexal lesions and ovarian implants in CT images
Jun 25, 2024



Abstract:Two self-supervised pretrained transformer-based segmentation models (SMIT and Swin UNETR) fine-tuned on a dataset of ovarian cancer CT images provided reasonably accurate delineations of the tumors in an independent test dataset. Tumors in the adnexa were segmented more accurately by both transformers (SMIT and Swin UNETR) than the omental implants. AI-assisted labeling performed on 72 out of 245 omental implants resulted in smaller manual editing effort of 39.55 mm compared to full manual correction of partial labels of 106.49 mm and resulted in overall improved accuracy performance. Both SMIT and Swin UNETR did not generate any false detection of omental metastases in the urinary bladder and relatively few false detections in the small bowel, with 2.16 cc on average for SMIT and 7.37 cc for Swin UNETR respectively.
Self-supervised learning improves robustness of deep learning lung tumor segmentation to CT imaging differences
May 14, 2024



Abstract:Self-supervised learning (SSL) is an approach to extract useful feature representations from unlabeled data, and enable fine-tuning on downstream tasks with limited labeled examples. Self-pretraining is a SSL approach that uses the curated task dataset for both pretraining the networks and fine-tuning them. Availability of large, diverse, and uncurated public medical image sets provides the opportunity to apply SSL in the "wild" and potentially extract features robust to imaging variations. However, the benefit of wild- vs self-pretraining has not been studied for medical image analysis. In this paper, we compare robustness of wild versus self-pretrained transformer (vision transformer [ViT] and hierarchical shifted window [Swin]) models to computed tomography (CT) imaging differences for non-small cell lung cancer (NSCLC) segmentation. Wild-pretrained Swin models outperformed self-pretrained Swin for the various imaging acquisitions. ViT resulted in similar accuracy for both wild- and self-pretrained models. Masked image prediction pretext task that forces networks to learn the local structure resulted in higher accuracy compared to contrastive task that models global image information. Wild-pretrained models resulted in higher feature reuse at the lower level layers and feature differentiation close to output layer after fine-tuning. Hence, we conclude: Wild-pretrained networks were more robust to analyzed CT imaging differences for lung tumor segmentation than self-pretrained methods. Swin architecture benefited from such pretraining more than ViT.
Deep learning classifier of locally advanced rectal cancer treatment response from endoscopy images
May 06, 2024



Abstract:We developed a deep learning classifier of rectal cancer response (tumor vs. no-tumor) to total neoadjuvant treatment (TNT) from endoscopic images acquired before, during, and following TNT. We further evaluated the network's ability in a near out-of-distribution (OOD) problem to identify local regrowth (LR) from follow-up endoscopy images acquired several months to years after completing TNT. We addressed endoscopic image variability by using optimal mass transport-based image harmonization. We evaluated multiple training regularization schemes to study the ResNet-50 network's in-distribution and near-OOD generalization ability. Test time augmentation resulted in the most considerable accuracy improvement. Image harmonization resulted in slight accuracy improvement for the near-OOD cases. Our results suggest that off-the-shelf deep learning classifiers can detect rectal cancer from endoscopic images at various stages of therapy for surveillance.
Trustworthiness of Pretrained Transformers for Lung Cancer Segmentation
Mar 19, 2024Abstract:We assessed the trustworthiness of two self-supervision pretrained transformer models, Swin UNETR and SMIT, for fine-tuned lung (LC) tumor segmentation using 670 CT and MRI scans. We measured segmentation accuracy on two public 3D-CT datasets, robustness on CT scans of patients with COVID-19, CT scans of patients with ovarian cancer and T2-weighted MRI of men with prostate cancer, and zero-shot generalization of LC for T2-weighted MRIs. Both models demonstrated high accuracy on in-distribution data (Dice 0.80 for SMIT and 0.78 for Swin UNETR). SMIT showed similar near-out-of-distribution performance on CT scans (AUROC 89.85% vs. 89.19%) but significantly better far-out-of-distribution accuracy on CT (AUROC 97.2% vs. 87.1%) and MRI (92.15% vs. 73.8%). SMIT outperformed Swin UNETR in zero-shot segmentation on MRI (Dice 0.78 vs. 0.69). We expect these findings to guide the safe development and deployment of current and future pretrained models in routine clinical use.
Semantic Segmentation with Active Semi-Supervised Representation Learning
Oct 16, 2022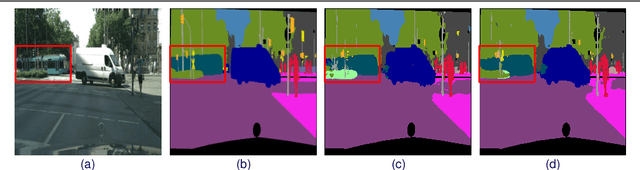
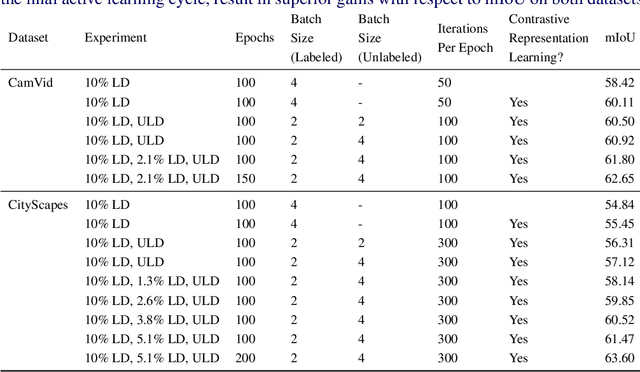
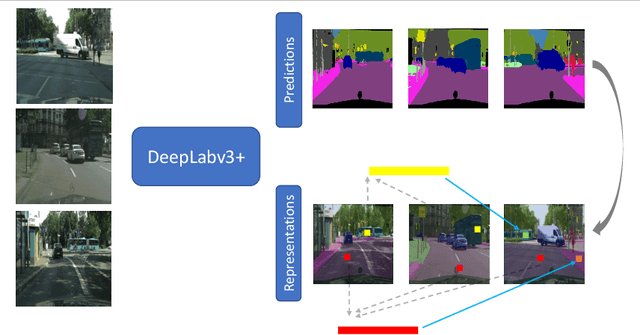
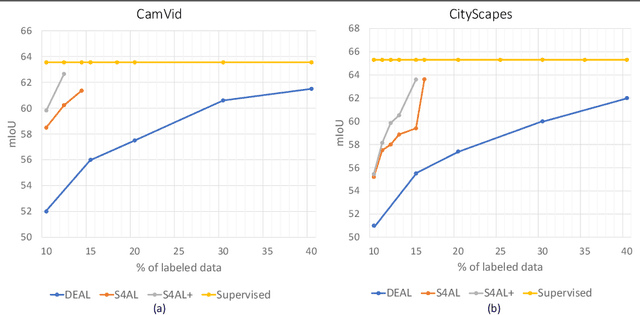
Abstract:Obtaining human per-pixel labels for semantic segmentation is incredibly laborious, often making labeled dataset construction prohibitively expensive. Here, we endeavor to overcome this problem with a novel algorithm that combines semi-supervised and active learning, resulting in the ability to train an effective semantic segmentation algorithm with significantly lesser labeled data. To do this, we extend the prior state-of-the-art S4AL algorithm by replacing its mean teacher approach for semi-supervised learning with a self-training approach that improves learning with noisy labels. We further boost the neural network's ability to query useful data by adding a contrastive learning head, which leads to better understanding of the objects in the scene, and hence, better queries for active learning. We evaluate our method on CamVid and CityScapes datasets, the de-facto standards for active learning for semantic segmentation. We achieve more than 95% of the network's performance on CamVid and CityScapes datasets, utilizing only 12.1% and 15.1% of the labeled data, respectively. We also benchmark our method across existing stand-alone semi-supervised learning methods on the CityScapes dataset and achieve superior performance without any bells or whistles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge