Image To Image Translation
Image-to-image translation is the process of converting an image from one domain to another using deep learning techniques.
Papers and Code
Augment to Augment: Diverse Augmentations Enable Competitive Ultra-Low-Field MRI Enhancement
Nov 12, 2025Ultra-low-field (ULF) MRI promises broader accessibility but suffers from low signal-to-noise ratio (SNR), reduced spatial resolution, and contrasts that deviate from high-field standards. Image-to-image translation can map ULF images to a high-field appearance, yet efficacy is limited by scarce paired training data. Working within the ULF-EnC challenge constraints (50 paired 3D volumes; no external data), we study how task-adapted data augmentations impact a standard deep model for ULF image enhancement. We show that strong, diverse augmentations, including auxiliary tasks on high-field data, substantially improve fidelity. Our submission ranked third by brain-masked SSIM on the public validation leaderboard and fourth by the official score on the final test leaderboard. Code is available at https://github.com/fzimmermann89/low-field-enhancement.
Electromagnetic Quantitative Inversion for Translationally Moving Targets via Phase Correlation Registration of Back-Projection Images
Nov 19, 2025A novel electromagnetic quantitative inversion scheme for translationally moving targets via phase correlation registration of back-projection (BP) images is proposed. Based on a time division multiplexing multiple-input multiple-output (TDM-MIMO) radar architecture, the scheme first achieves high-precision relative positioning of the target, then applies relative motion compensation to perform iterative inversion on multi-cycle MIMO measurement data, thereby reconstructing the target's electromagnetic parameters. As a general framework compatible with other mainstream inversion algorithms, we exemplify our approach by incorporating the classical cross-correlated contrast source inversion (CC-CSI) into iterative optimization step of the scheme, resulting in a new algorithm termed RMC-CC-CSI. Numerical and experimental results demonstrate that RMC-CC-CSI offers accelerated convergence, enhanced reconstruction fidelity, and improved noise immunity over conventional CC-CSI for stationary targets despite increased computational cost.
DINOv3-Guided Cross Fusion Framework for Semantic-aware CT generation from MRI and CBCT
Nov 15, 2025



Generating synthetic CT images from CBCT or MRI has a potential for efficient radiation dose planning and adaptive radiotherapy. However, existing CNN-based models lack global semantic understanding, while Transformers often overfit small medical datasets due to high model capacity and weak inductive bias. To address these limitations, we propose a DINOv3-Guided Cross Fusion (DGCF) framework that integrates a frozen self-supervised DINOv3 Transformer with a trainable CNN encoder-decoder. It hierarchically fuses global representation of Transformer and local features of CNN via a learnable cross fusion module, achieving balanced local appearance and contextual representation. Furthermore, we introduce a Multi-Level DINOv3 Perceptual (MLDP) loss that encourages semantic similarity between synthetic CT and the ground truth in DINOv3's feature space. Experiments on the SynthRAD2023 pelvic dataset demonstrate that DGCF achieved state-of-the-art performance in terms of MS-SSIM, PSNR and segmentation-based metrics on both MRI$\rightarrow$CT and CBCT$\rightarrow$CT translation tasks. To the best of our knowledge, this is the first work to employ DINOv3 representations for medical image translation, highlighting the potential of self-supervised Transformer guidance for semantic-aware CT synthesis. The code is available at https://github.com/HiLab-git/DGCF.
Lost in Translation, Found in Embeddings: Sign Language Translation and Alignment
Dec 08, 2025


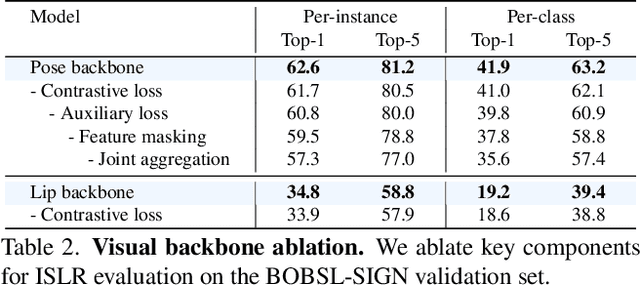
Our aim is to develop a unified model for sign language understanding, that performs sign language translation (SLT) and sign-subtitle alignment (SSA). Together, these two tasks enable the conversion of continuous signing videos into spoken language text and also the temporal alignment of signing with subtitles -- both essential for practical communication, large-scale corpus construction, and educational applications. To achieve this, our approach is built upon three components: (i) a lightweight visual backbone that captures manual and non-manual cues from human keypoints and lip-region images while preserving signer privacy; (ii) a Sliding Perceiver mapping network that aggregates consecutive visual features into word-level embeddings to bridge the vision-text gap; and (iii) a multi-task scalable training strategy that jointly optimises SLT and SSA, reinforcing both linguistic and temporal alignment. To promote cross-linguistic generalisation, we pretrain our model on large-scale sign-text corpora covering British Sign Language (BSL) and American Sign Language (ASL) from the BOBSL and YouTube-SL-25 datasets. With this multilingual pretraining and strong model design, we achieve state-of-the-art results on the challenging BOBSL (BSL) dataset for both SLT and SSA. Our model also demonstrates robust zero-shot generalisation and finetuned SLT performance on How2Sign (ASL), highlighting the potential of scalable translation across different sign languages.
Scene-agnostic Hierarchical Bimanual Task Planning via Visual Affordance Reasoning
Dec 10, 2025Embodied agents operating in open environments must translate high-level instructions into grounded, executable behaviors, often requiring coordinated use of both hands. While recent foundation models offer strong semantic reasoning, existing robotic task planners remain predominantly unimanual and fail to address the spatial, geometric, and coordination challenges inherent to bimanual manipulation in scene-agnostic settings. We present a unified framework for scene-agnostic bimanual task planning that bridges high-level reasoning with 3D-grounded two-handed execution. Our approach integrates three key modules. Visual Point Grounding (VPG) analyzes a single scene image to detect relevant objects and generate world-aligned interaction points. Bimanual Subgoal Planner (BSP) reasons over spatial adjacency and cross-object accessibility to produce compact, motion-neutralized subgoals that exploit opportunities for coordinated two-handed actions. Interaction-Point-Driven Bimanual Prompting (IPBP) binds these subgoals to a structured skill library, instantiating synchronized unimanual or bimanual action sequences that satisfy hand-state and affordance constraints. Together, these modules enable agents to plan semantically meaningful, physically feasible, and parallelizable two-handed behaviors in cluttered, previously unseen scenes. Experiments show that it produces coherent, feasible, and compact two-handed plans, and generalizes to cluttered scenes without retraining, demonstrating robust scene-agnostic affordance reasoning for bimanual tasks.
Regional Attention-Enhanced Swin Transformer for Clinically Relevant Medical Image Captioning
Nov 13, 2025Automated medical image captioning translates complex radiological images into diagnostic narratives that can support reporting workflows. We present a Swin-BART encoder-decoder system with a lightweight regional attention module that amplifies diagnostically salient regions before cross-attention. Trained and evaluated on ROCO, our model achieves state-of-the-art semantic fidelity while remaining compact and interpretable. We report results as mean$\pm$std over three seeds and include $95\%$ confidence intervals. Compared with baselines, our approach improves ROUGE (proposed 0.603, ResNet-CNN 0.356, BLIP2-OPT 0.255) and BERTScore (proposed 0.807, BLIP2-OPT 0.645, ResNet-CNN 0.623), with competitive BLEU, CIDEr, and METEOR. We further provide ablations (regional attention on/off and token-count sweep), per-modality analysis (CT/MRI/X-ray), paired significance tests, and qualitative heatmaps that visualize the regions driving each description. Decoding uses beam search (beam size $=4$), length penalty $=1.1$, $no\_repeat\_ngram\_size$ $=3$, and max length $=128$. The proposed design yields accurate, clinically phrased captions and transparent regional attributions, supporting safe research use with a human in the loop.
DIST-CLIP: Arbitrary Metadata and Image Guided MRI Harmonization via Disentangled Anatomy-Contrast Representations
Dec 08, 2025Deep learning holds immense promise for transforming medical image analysis, yet its clinical generalization remains profoundly limited. A major barrier is data heterogeneity. This is particularly true in Magnetic Resonance Imaging, where scanner hardware differences, diverse acquisition protocols, and varying sequence parameters introduce substantial domain shifts that obscure underlying biological signals. Data harmonization methods aim to reduce these instrumental and acquisition variability, but existing approaches remain insufficient. When applied to imaging data, image-based harmonization approaches are often restricted by the need for target images, while existing text-guided methods rely on simplistic labels that fail to capture complex acquisition details or are typically restricted to datasets with limited variability, failing to capture the heterogeneity of real-world clinical environments. To address these limitations, we propose DIST-CLIP (Disentangled Style Transfer with CLIP Guidance), a unified framework for MRI harmonization that flexibly uses either target images or DICOM metadata for guidance. Our framework explicitly disentangles anatomical content from image contrast, with the contrast representations being extracted using pre-trained CLIP encoders. These contrast embeddings are then integrated into the anatomical content via a novel Adaptive Style Transfer module. We trained and evaluated DIST-CLIP on diverse real-world clinical datasets, and showed significant improvements in performance when compared against state-of-the-art methods in both style translation fidelity and anatomical preservation, offering a flexible solution for style transfer and standardizing MRI data. Our code and weights will be made publicly available upon publication.
DT-NVS: Diffusion Transformers for Novel View Synthesis
Nov 11, 2025Generating novel views of a natural scene, e.g., every-day scenes both indoors and outdoors, from a single view is an under-explored problem, even though it is an organic extension to the object-centric novel view synthesis. Existing diffusion-based approaches focus rather on small camera movements in real scenes or only consider unnatural object-centric scenes, limiting their potential applications in real-world settings. In this paper we move away from these constrained regimes and propose a 3D diffusion model trained with image-only losses on a large-scale dataset of real-world, multi-category, unaligned, and casually acquired videos of everyday scenes. We propose DT-NVS, a 3D-aware diffusion model for generalized novel view synthesis that exploits a transformer-based architecture backbone. We make significant contributions to transformer and self-attention architectures to translate images to 3d representations, and novel camera conditioning strategies to allow training on real-world unaligned datasets. In addition, we introduce a novel training paradigm swapping the role of reference frame between the conditioning image and the sampled noisy input. We evaluate our approach on the 3D task of generalized novel view synthesis from a single input image and show improvements over state-of-the-art 3D aware diffusion models and deterministic approaches, while generating diverse outputs.
MVP: Multiple View Prediction Improves GUI Grounding
Dec 09, 2025



GUI grounding, which translates natural language instructions into precise pixel coordinates, is essential for developing practical GUI agents. However, we observe that existing grounding models exhibit significant coordinate prediction instability, minor visual perturbations (e.g. cropping a few pixels) can drastically alter predictions, flipping results between correct and incorrect. This instability severely undermines model performance, especially for samples with high-resolution and small UI elements. To address this issue, we propose Multi-View Prediction (MVP), a training-free framework that enhances grounding performance through multi-view inference. Our key insight is that while single-view predictions may be unstable, aggregating predictions from multiple carefully cropped views can effectively distinguish correct coordinates from outliers. MVP comprises two components: (1) Attention-Guided View Proposal, which derives diverse views guided by instruction-to-image attention scores, and (2) Multi-Coordinates Clustering, which ensembles predictions by selecting the centroid of the densest spatial cluster. Extensive experiments demonstrate MVP's effectiveness across various models and benchmarks. Notably, on ScreenSpot-Pro, MVP boosts UI-TARS-1.5-7B to 56.1%, GTA1-7B to 61.7%, Qwen3VL-8B-Instruct to 65.3%, and Qwen3VL-32B-Instruct to 74.0%. The code is available at https://github.com/ZJUSCL/MVP.
TraceTrans: Translation and Spatial Tracing for Surgical Prediction
Oct 25, 2025Image-to-image translation models have achieved notable success in converting images across visual domains and are increasingly used for medical tasks such as predicting post-operative outcomes and modeling disease progression. However, most existing methods primarily aim to match the target distribution and often neglect spatial correspondences between the source and translated images. This limitation can lead to structural inconsistencies and hallucinations, undermining the reliability and interpretability of the predictions. These challenges are accentuated in clinical applications by the stringent requirement for anatomical accuracy. In this work, we present TraceTrans, a novel deformable image translation model designed for post-operative prediction that generates images aligned with the target distribution while explicitly revealing spatial correspondences with the pre-operative input. The framework employs an encoder for feature extraction and dual decoders for predicting spatial deformations and synthesizing the translated image. The predicted deformation field imposes spatial constraints on the generated output, ensuring anatomical consistency with the source. Extensive experiments on medical cosmetology and brain MRI datasets demonstrate that TraceTrans delivers accurate and interpretable post-operative predictions, highlighting its potential for reliable clinical deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge