Zhuangzhuang Chen
Boosting RL-Based Visual Reasoning with Selective Adversarial Entropy Intervention
Dec 11, 2025



Abstract:Recently, reinforcement learning (RL) has become a common choice in enhancing the reasoning capabilities of vision-language models (VLMs). Considering existing RL-based finetuning methods, entropy intervention turns out to be an effective way to benefit exploratory ability, thereby improving policy performance. Notably, most existing studies intervene in entropy by simply controlling the update of specific tokens during policy optimization of RL. They ignore the entropy intervention during the RL sampling that can boost the performance of GRPO by improving the diversity of responses. In this paper, we propose Selective-adversarial Entropy Intervention, namely SaEI, which enhances policy entropy by distorting the visual input with the token-selective adversarial objective coming from the entropy of sampled responses. Specifically, we first propose entropy-guided adversarial sampling (EgAS) that formulates the entropy of sampled responses as an adversarial objective. Then, the corresponding adversarial gradient can be used to attack the visual input for producing adversarial samples, allowing the policy model to explore a larger answer space during RL sampling. Then, we propose token-selective entropy computation (TsEC) to maximize the effectiveness of adversarial attack in EgAS without distorting factual knowledge within VLMs. Extensive experiments on both in-domain and out-of-domain datasets show that our proposed method can greatly improve policy exploration via entropy intervention, to boost reasoning capabilities. Code will be released once the paper is accepted.
Leveraging Segment Anything Model for Source-Free Domain Adaptation via Dual Feature Guided Auto-Prompting
May 14, 2025Abstract:Source-free domain adaptation (SFDA) for segmentation aims at adapting a model trained in the source domain to perform well in the target domain with only the source model and unlabeled target data.Inspired by the recent success of Segment Anything Model (SAM) which exhibits the generality of segmenting images of various modalities and in different domains given human-annotated prompts like bounding boxes or points, we for the first time explore the potentials of Segment Anything Model for SFDA via automatedly finding an accurate bounding box prompt. We find that the bounding boxes directly generated with existing SFDA approaches are defective due to the domain gap.To tackle this issue, we propose a novel Dual Feature Guided (DFG) auto-prompting approach to search for the box prompt. Specifically, the source model is first trained in a feature aggregation phase, which not only preliminarily adapts the source model to the target domain but also builds a feature distribution well-prepared for box prompt search. In the second phase, based on two feature distribution observations, we gradually expand the box prompt with the guidance of the target model feature and the SAM feature to handle the class-wise clustered target features and the class-wise dispersed target features, respectively. To remove the potentially enlarged false positive regions caused by the over-confident prediction of the target model, the refined pseudo-labels produced by SAM are further postprocessed based on connectivity analysis. Experiments on 3D and 2D datasets indicate that our approach yields superior performance compared to conventional methods. Code is available at https://github.com/xmed-lab/DFG.
MuTri: Multi-view Tri-alignment for OCT to OCTA 3D Image Translation
Apr 02, 2025



Abstract:Optical coherence tomography angiography (OCTA) shows its great importance in imaging microvascular networks by providing accurate 3D imaging of blood vessels, but it relies upon specialized sensors and expensive devices. For this reason, previous works show the potential to translate the readily available 3D Optical Coherence Tomography (OCT) images into 3D OCTA images. However, existing OCTA translation methods directly learn the mapping from the OCT domain to the OCTA domain in continuous and infinite space with guidance from only a single view, i.e., the OCTA project map, resulting in suboptimal results. To this end, we propose the multi-view Tri-alignment framework for OCT to OCTA 3D image translation in discrete and finite space, named MuTri. In the first stage, we pre-train two vector-quantized variational auto-encoder (VQ- VAE) by reconstructing 3D OCT and 3D OCTA data, providing semantic prior for subsequent multi-view guidances. In the second stage, our multi-view tri-alignment facilitates another VQVAE model to learn the mapping from the OCT domain to the OCTA domain in discrete and finite space. Specifically, a contrastive-inspired semantic alignment is proposed to maximize the mutual information with the pre-trained models from OCT and OCTA views, to facilitate codebook learning. Meanwhile, a vessel structure alignment is proposed to minimize the structure discrepancy with the pre-trained models from the OCTA project map view, benefiting from learning the detailed vessel structure information. We also collect the first large-scale dataset, namely, OCTA2024, which contains a pair of OCT and OCTA volumes from 846 subjects.
Task-Specific Knowledge Distillation from the Vision Foundation Model for Enhanced Medical Image Segmentation
Mar 10, 2025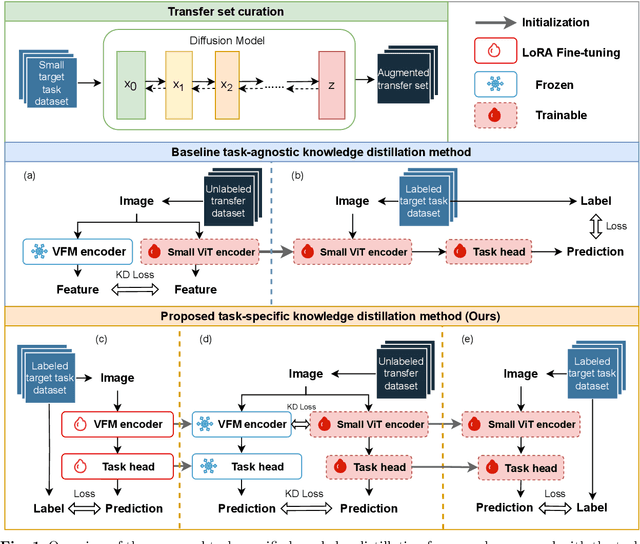
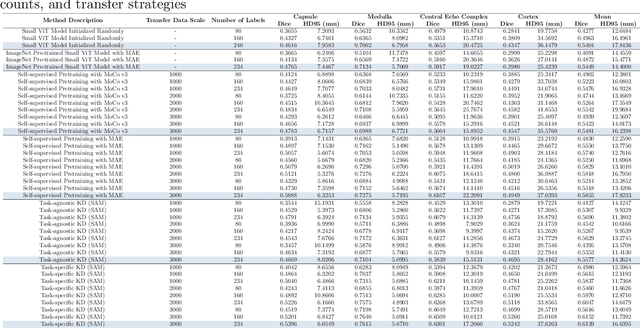
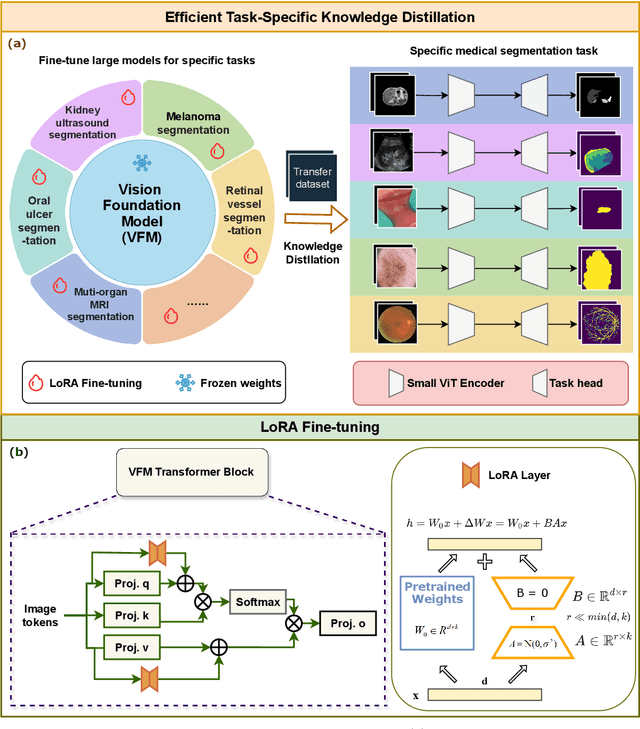
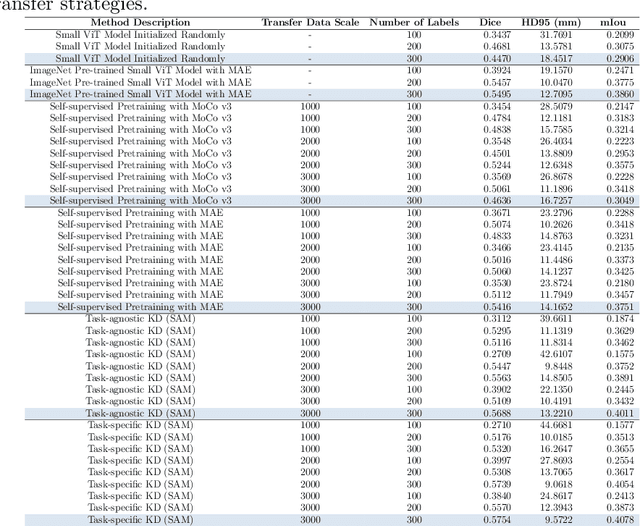
Abstract:Large-scale pre-trained models, such as Vision Foundation Models (VFMs), have demonstrated impressive performance across various downstream tasks by transferring generalized knowledge, especially when target data is limited. However, their high computational cost and the domain gap between natural and medical images limit their practical application in medical segmentation tasks. Motivated by this, we pose the following important question: "How can we effectively utilize the knowledge of large pre-trained VFMs to train a small, task-specific model for medical image segmentation when training data is limited?" To address this problem, we propose a novel and generalizable task-specific knowledge distillation framework. Our method fine-tunes the VFM on the target segmentation task to capture task-specific features before distilling the knowledge to smaller models, leveraging Low-Rank Adaptation (LoRA) to reduce the computational cost of fine-tuning. Additionally, we incorporate synthetic data generated by diffusion models to augment the transfer set, enhancing model performance in data-limited scenarios. Experimental results across five medical image datasets demonstrate that our method consistently outperforms task-agnostic knowledge distillation and self-supervised pretraining approaches like MoCo v3 and Masked Autoencoders (MAE). For example, on the KidneyUS dataset, our method achieved a 28% higher Dice score than task-agnostic KD using 80 labeled samples for fine-tuning. On the CHAOS dataset, it achieved an 11% improvement over MAE with 100 labeled samples. These results underscore the potential of task-specific knowledge distillation to train accurate, efficient models for medical image segmentation in data-constrained settings.
Semantic Prior Distillation with Vision Foundation Model for Enhanced Rapid Bone Scintigraphy Image Restoration
Mar 04, 2025



Abstract:Rapid bone scintigraphy is an essential tool for diagnosing skeletal diseases and tumor metastasis in pediatric patients, as it reduces scan time and minimizes patient discomfort. However, rapid scans often result in poor image quality, potentially affecting diagnosis due to reduced resolution and detail, which make it challenging to identify and evaluate finer anatomical structures. To address this issue, we propose the first application of SAM-based semantic priors for medical image restoration, leveraging the Segment Anything Model (SAM) to enhance rapid bone scintigraphy images in pediatric populations. Our method comprises two cascaded networks, $f^{IR1}$ and $f^{IR2}$, augmented by three key modules: a Semantic Prior Integration (SPI) module, a Semantic Knowledge Distillation (SKD) module, and a Semantic Consistency Module (SCM). The SPI and SKD modules incorporate domain-specific semantic information from a fine-tuned SAM, while the SCM maintains consistent semantic feature representation throughout the cascaded networks. In addition, we will release a novel Rapid Bone Scintigraphy dataset called RBS, the first dataset dedicated to rapid bone scintigraphy image restoration in pediatric patients. RBS consists of 137 pediatric patients aged between 0.5 and 16 years who underwent both standard and rapid bone scans. The dataset includes scans performed at 20 cm/min (standard) and 40 cm/min (rapid), representing a $2\times$ acceleration. We conducted extensive experiments on both the publicly available endoscopic dataset and RBS. The results demonstrate that our method outperforms all existing methods across various metrics, including PSNR, SSIM, FID, and LPIPS.
Shift-ConvNets: Small Convolutional Kernel with Large Kernel Effects
Jan 23, 2024Abstract:Recent studies reveal that the remarkable performance of Vision transformers (ViTs) benefits from large receptive fields. For this reason, the large convolutional kernel design becomes an ideal solution to make Convolutional Neural Networks (CNNs) great again. However, the typical large convolutional kernels turn out to be hardware-unfriendly operators, resulting in discount compatibility of various hardware platforms. Thus, it is unwise to simply enlarge the convolutional kernel size. In this paper, we reveal that small convolutional kernels and convolution operations can achieve the closing effects of large kernel sizes. Then, we propose a shift-wise operator that ensures the CNNs capture long-range dependencies with the help of the sparse mechanism, while remaining hardware-friendly. Experimental results show that our shift-wise operator significantly improves the accuracy of a regular CNN while markedly reducing computational requirements. On the ImageNet-1k, our shift-wise enhanced CNN model outperforms the state-of-the-art models. Code & models at https://github.com/lidc54/shift-wiseConv.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge