Yuezhi Cai
AdaptCLIP: Adapting CLIP for Universal Visual Anomaly Detection
May 15, 2025Abstract:Universal visual anomaly detection aims to identify anomalies from novel or unseen vision domains without additional fine-tuning, which is critical in open scenarios. Recent studies have demonstrated that pre-trained vision-language models like CLIP exhibit strong generalization with just zero or a few normal images. However, existing methods struggle with designing prompt templates, complex token interactions, or requiring additional fine-tuning, resulting in limited flexibility. In this work, we present a simple yet effective method called AdaptCLIP based on two key insights. First, adaptive visual and textual representations should be learned alternately rather than jointly. Second, comparative learning between query and normal image prompt should incorporate both contextual and aligned residual features, rather than relying solely on residual features. AdaptCLIP treats CLIP models as a foundational service, adding only three simple adapters, visual adapter, textual adapter, and prompt-query adapter, at its input or output ends. AdaptCLIP supports zero-/few-shot generalization across domains and possesses a training-free manner on target domains once trained on a base dataset. AdaptCLIP achieves state-of-the-art performance on 12 anomaly detection benchmarks from industrial and medical domains, significantly outperforming existing competitive methods. We will make the code and model of AdaptCLIP available at https://github.com/gaobb/AdaptCLIP.
Explaining COVID-19 and Thoracic Pathology Model Predictions by Identifying Informative Input Features
Apr 01, 2021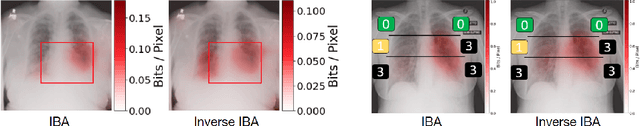
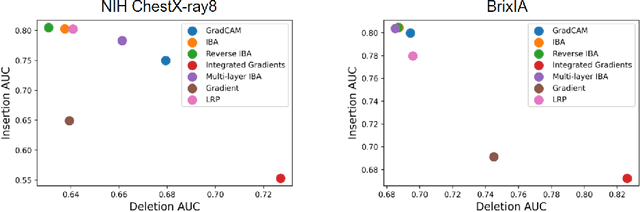
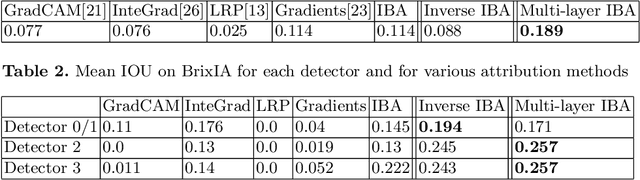
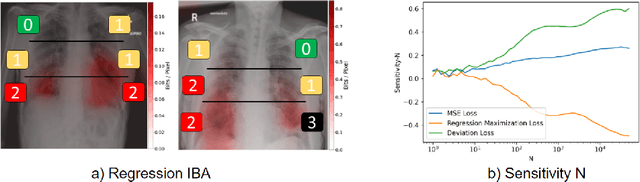
Abstract:Neural networks have demonstrated remarkable performance in classification and regression tasks on chest X-rays. In order to establish trust in the clinical routine, the networks' prediction mechanism needs to be interpretable. One principal approach to interpretation is feature attribution. Feature attribution methods identify the importance of input features for the output prediction. Building on Information Bottleneck Attribution (IBA) method, for each prediction we identify the chest X-ray regions that have high mutual information with the network's output. Original IBA identifies input regions that have sufficient predictive information. We propose Inverse IBA to identify all informative regions. Thus all predictive cues for pathologies are highlighted on the X-rays, a desirable property for chest X-ray diagnosis. Moreover, we propose Regression IBA for explaining regression models. Using Regression IBA we observe that a model trained on cumulative severity score labels implicitly learns the severity of different X-ray regions. Finally, we propose Multi-layer IBA to generate higher resolution and more detailed attribution/saliency maps. We evaluate our methods using both human-centric (ground-truth-based) interpretability metrics, and human-independent feature importance metrics on NIH Chest X-ray8 and BrixIA datasets. The Code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge