Weijie Sun
Bohrium + SciMaster: Building the Infrastructure and Ecosystem for Agentic Science at Scale
Dec 23, 2025



Abstract:AI agents are emerging as a practical way to run multi-step scientific workflows that interleave reasoning with tool use and verification, pointing to a shift from isolated AI-assisted steps toward \emph{agentic science at scale}. This shift is increasingly feasible, as scientific tools and models can be invoked through stable interfaces and verified with recorded execution traces, and increasingly necessary, as AI accelerates scientific output and stresses the peer-review and publication pipeline, raising the bar for traceability and credible evaluation. However, scaling agentic science remains difficult: workflows are hard to observe and reproduce; many tools and laboratory systems are not agent-ready; execution is hard to trace and govern; and prototype AI Scientist systems are often bespoke, limiting reuse and systematic improvement from real workflow signals. We argue that scaling agentic science requires an infrastructure-and-ecosystem approach, instantiated in Bohrium+SciMaster. Bohrium acts as a managed, traceable hub for AI4S assets -- akin to a HuggingFace of AI for Science -- that turns diverse scientific data, software, compute, and laboratory systems into agent-ready capabilities. SciMaster orchestrates these capabilities into long-horizon scientific workflows, on which scientific agents can be composed and executed. Between infrastructure and orchestration, a \emph{scientific intelligence substrate} organizes reusable models, knowledge, and components into executable building blocks for workflow reasoning and action, enabling composition, auditability, and improvement through use. We demonstrate this stack with eleven representative master agents in real workflows, achieving orders-of-magnitude reductions in end-to-end scientific cycle time and generating execution-grounded signals from real workloads at multi-million scale.
Inverse Knowledge Search over Verifiable Reasoning: Synthesizing a Scientific Encyclopedia from a Long Chains-of-Thought Knowledge Base
Oct 30, 2025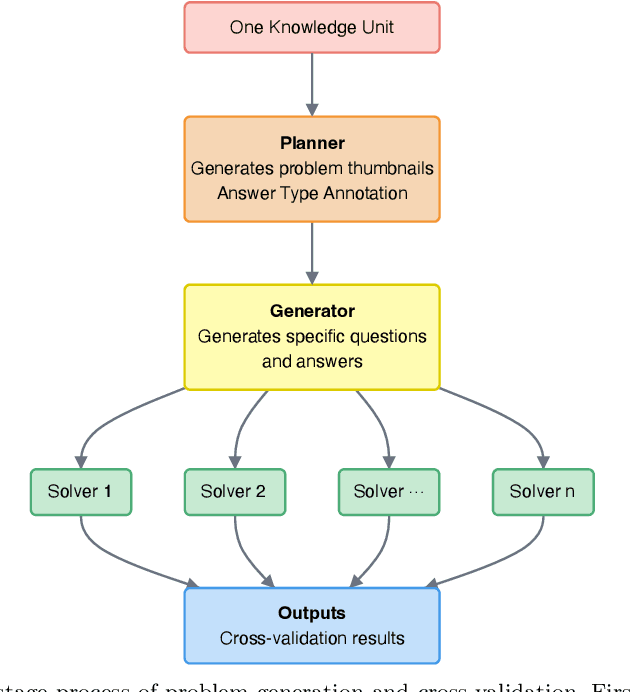
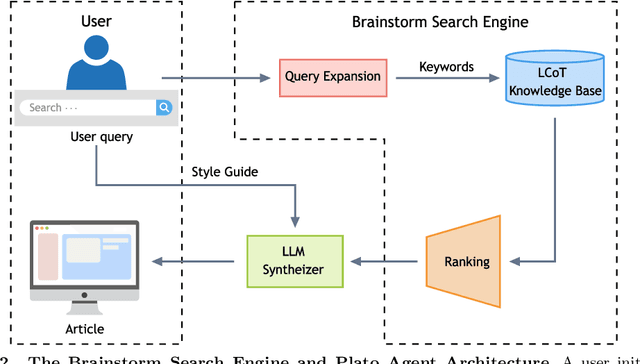
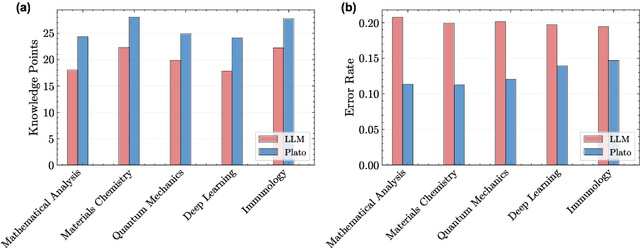
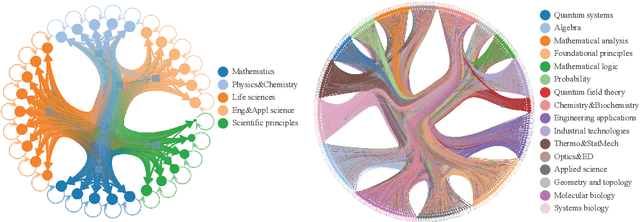
Abstract:Most scientific materials compress reasoning, presenting conclusions while omitting the derivational chains that justify them. This compression hinders verification by lacking explicit, step-wise justifications and inhibits cross-domain links by collapsing the very pathways that establish the logical and causal connections between concepts. We introduce a scalable framework that decompresses scientific reasoning, constructing a verifiable Long Chain-of-Thought (LCoT) knowledge base and projecting it into an emergent encyclopedia, SciencePedia. Our pipeline operationalizes an endpoint-driven, reductionist strategy: a Socratic agent, guided by a curriculum of around 200 courses, generates approximately 3 million first-principles questions. To ensure high fidelity, multiple independent solver models generate LCoTs, which are then rigorously filtered by prompt sanitization and cross-model answer consensus, retaining only those with verifiable endpoints. This verified corpus powers the Brainstorm Search Engine, which performs inverse knowledge search -- retrieving diverse, first-principles derivations that culminate in a target concept. This engine, in turn, feeds the Plato synthesizer, which narrates these verified chains into coherent articles. The initial SciencePedia comprises approximately 200,000 fine-grained entries spanning mathematics, physics, chemistry, biology, engineering, and computation. In evaluations across six disciplines, Plato-synthesized articles (conditioned on retrieved LCoTs) exhibit substantially higher knowledge-point density and significantly lower factual error rates than an equally-prompted baseline without retrieval (as judged by an external LLM). Built on this verifiable LCoT knowledge base, this reasoning-centric approach enables trustworthy, cross-domain scientific synthesis at scale and establishes the foundation for an ever-expanding encyclopedia.
MENSA: A Multi-Event Network for Survival Analysis under Informative Censoring
Sep 10, 2024



Abstract:Given an instance, a multi-event survival model predicts the time until that instance experiences each of several different events. These events are not mutually exclusive and there are often statistical dependencies between them. There are relatively few multi-event survival results, most focusing on producing a simple risk score, rather than the time-to-event itself. To overcome these issues, we introduce MENSA, a novel, deep learning approach for multi-event survival analysis that can jointly learn representations of the input covariates and the dependence structure between events. As a practical motivation for multi-event survival analysis, we consider the problem of predicting the time until a patient with amyotrophic lateral sclerosis (ALS) loses various physical functions, i.e., the ability to speak, swallow, write, or walk. When estimating when a patient is no longer able to swallow, our approach achieves an L1-Margin loss of 278.8 days, compared to 355.2 days when modeling each event separately. In addition, we also evaluate our approach in single-event and competing risk scenarios by modeling the censoring and event distributions as equal contributing factors in the optimization process, and show that our approach performs well across multiple benchmark datasets. The source code is available at: https://github.com/thecml/mensa
An Effective Meaningful Way to Evaluate Survival Models
Jun 01, 2023Abstract:One straightforward metric to evaluate a survival prediction model is based on the Mean Absolute Error (MAE) -- the average of the absolute difference between the time predicted by the model and the true event time, over all subjects. Unfortunately, this is challenging because, in practice, the test set includes (right) censored individuals, meaning we do not know when a censored individual actually experienced the event. In this paper, we explore various metrics to estimate MAE for survival datasets that include (many) censored individuals. Moreover, we introduce a novel and effective approach for generating realistic semi-synthetic survival datasets to facilitate the evaluation of metrics. Our findings, based on the analysis of the semi-synthetic datasets, reveal that our proposed metric (MAE using pseudo-observations) is able to rank models accurately based on their performance, and often closely matches the true MAE -- in particular, is better than several alternative methods.
Improving ECG-based COVID-19 diagnosis and mortality predictions using pre-pandemic medical records at population-scale
Nov 14, 2022



Abstract:Pandemic outbreaks such as COVID-19 occur unexpectedly, and need immediate action due to their potential devastating consequences on global health. Point-of-care routine assessments such as electrocardiogram (ECG), can be used to develop prediction models for identifying individuals at risk. However, there is often too little clinically-annotated medical data, especially in early phases of a pandemic, to develop accurate prediction models. In such situations, historical pre-pandemic health records can be utilized to estimate a preliminary model, which can then be fine-tuned based on limited available pandemic data. This study shows this approach -- pre-train deep learning models with pre-pandemic data -- can work effectively, by demonstrating substantial performance improvement over three different COVID-19 related diagnostic and prognostic prediction tasks. Similar transfer learning strategies can be useful for developing timely artificial intelligence solutions in future pandemic outbreaks.
ECG for high-throughput screening of multiple diseases: Proof-of-concept using multi-diagnosis deep learning from population-based datasets
Oct 06, 2022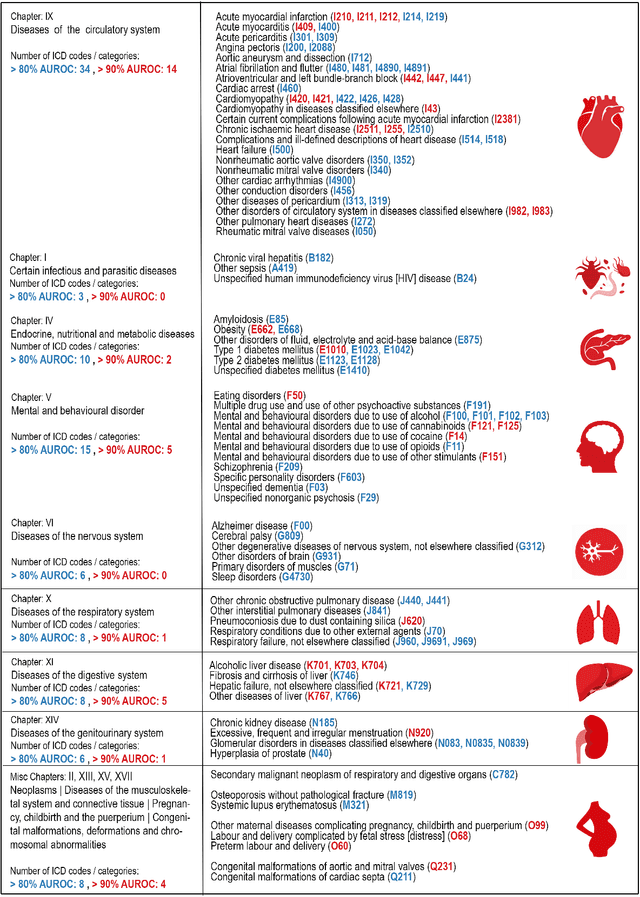
Abstract:Electrocardiogram (ECG) abnormalities are linked to cardiovascular diseases, but may also occur in other non-cardiovascular conditions such as mental, neurological, metabolic and infectious conditions. However, most of the recent success of deep learning (DL) based diagnostic predictions in selected patient cohorts have been limited to a small set of cardiac diseases. In this study, we use a population-based dataset of >250,000 patients with >1000 medical conditions and >2 million ECGs to identify a wide range of diseases that could be accurately diagnosed from the patient's first in-hospital ECG. Our DL models uncovered 128 diseases and 68 disease categories with strong discriminative performance.
Multilabel 12-Lead Electrocardiogram Classification Using Gradient Boosting Tree Ensemble
Oct 21, 2020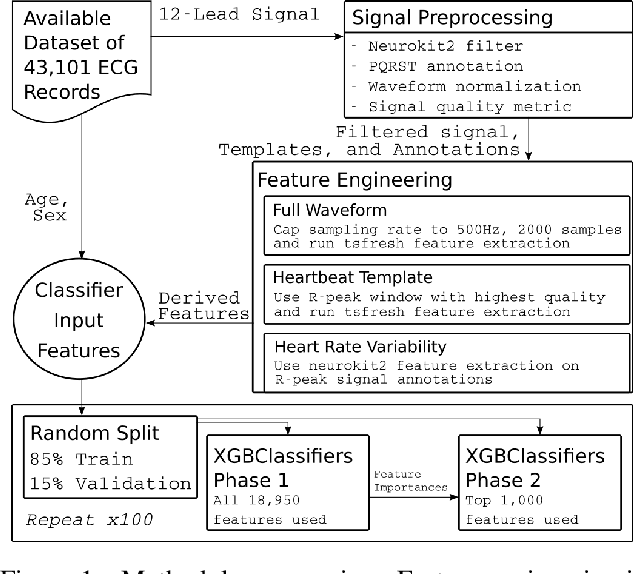
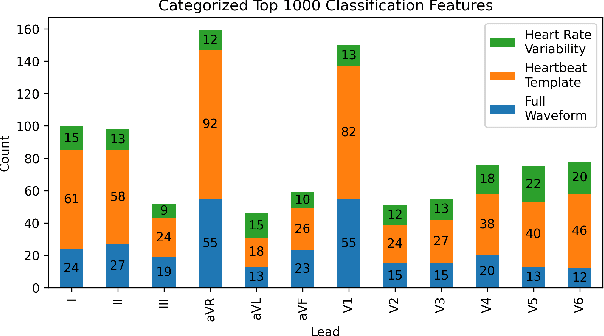
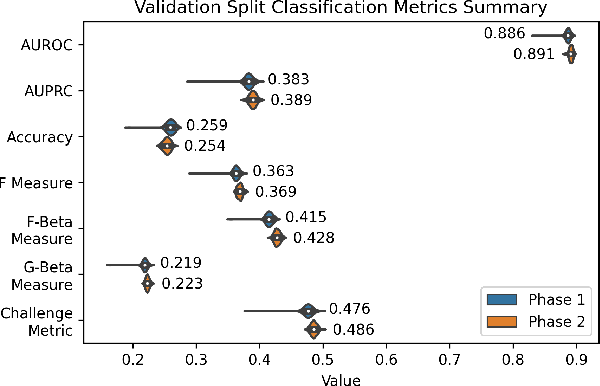
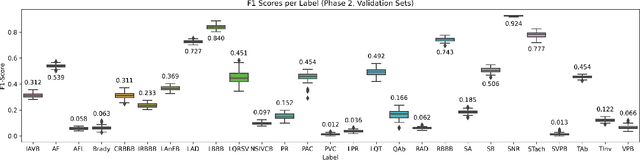
Abstract:The 12-lead electrocardiogram (ECG) is a commonly used tool for detecting cardiac abnormalities such as atrial fibrillation, blocks, and irregular complexes. For the PhysioNet/CinC 2020 Challenge, we built an algorithm using gradient boosted tree ensembles fitted on morphology and signal processing features to classify ECG diagnosis. For each lead, we derive features from heart rate variability, PQRST template shape, and the full signal waveform. We join the features of all 12 leads to fit an ensemble of gradient boosting decision trees to predict probabilities of ECG instances belonging to each class. We train a phase one set of feature importance determining models to isolate the top 1,000 most important features to use in our phase two diagnosis prediction models. We use repeated random sub-sampling by splitting our dataset of 43,101 records into 100 independent runs of 85:15 training/validation splits for our internal evaluation results. Our methodology generates us an official phase validation set score of 0.476 and test set score of -0.080 under the team name, CVC, placing us 36 out of 41 in the rankings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge