Tristan Naumann
Microsoft Research
Knowledge-Rich Self-Supervised Entity Linking
Dec 15, 2021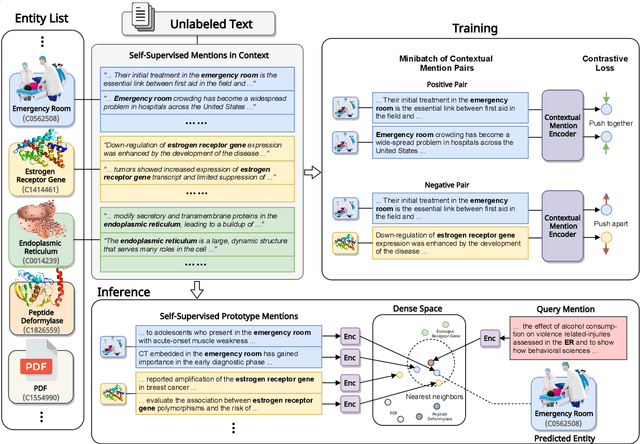
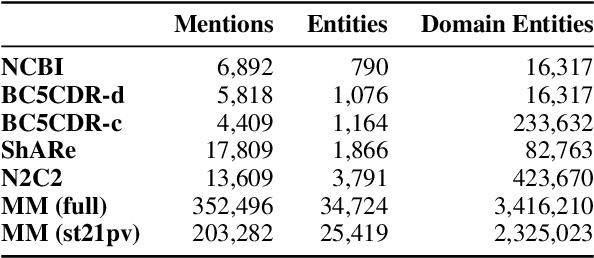
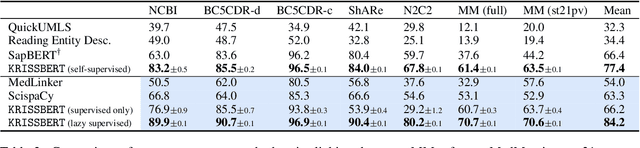
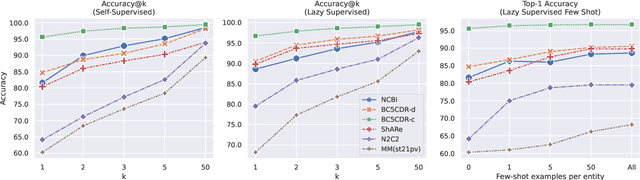
Abstract:Entity linking faces significant challenges, such as prolific variations and prevalent ambiguities, especially in high-value domains with myriad entities. Standard classification approaches suffer from the annotation bottleneck and cannot effectively handle unseen entities. Zero-shot entity linking has emerged as a promising direction for generalizing to new entities, but it still requires example gold entity mentions during training and canonical descriptions for all entities, both of which are rarely available outside of Wikipedia. In this paper, we explore Knowledge-RIch Self-Supervision ($\tt KRISS$) for entity linking, by leveraging readily available domain knowledge. In training, it generates self-supervised mention examples on unlabeled text using a domain ontology and trains a contextual encoder using contrastive learning. For inference, it samples self-supervised mentions as prototypes for each entity and conducts linking by mapping the test mention to the most similar prototype. Our approach subsumes zero-shot and few-shot methods, and can easily incorporate entity descriptions and gold mention labels if available. Using biomedicine as a case study, we conducted extensive experiments on seven standard datasets spanning biomedical literature and clinical notes. Without using any labeled information, our method produces $\tt KRISSBERT$, a universal entity linker for four million UMLS entities, which attains new state of the art, outperforming prior self-supervised methods by as much as over 20 absolute points in accuracy.
Fine-Tuning Large Neural Language Models for Biomedical Natural Language Processing
Dec 15, 2021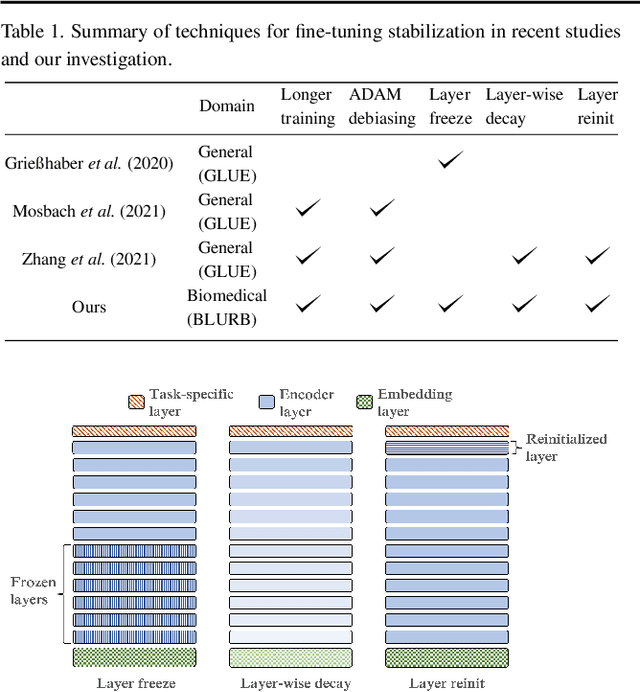
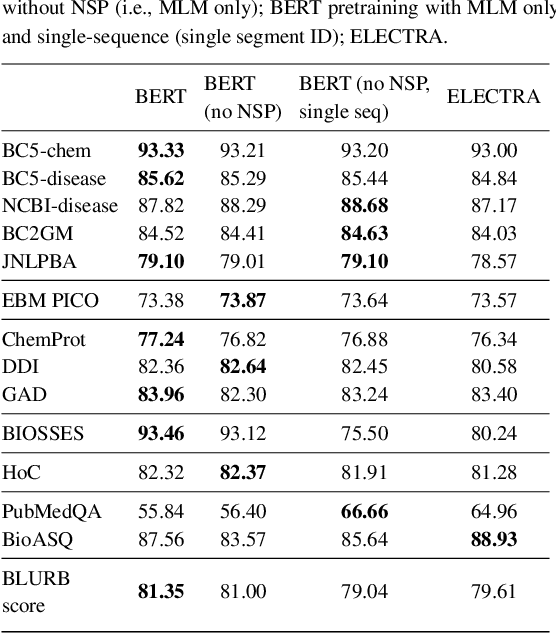
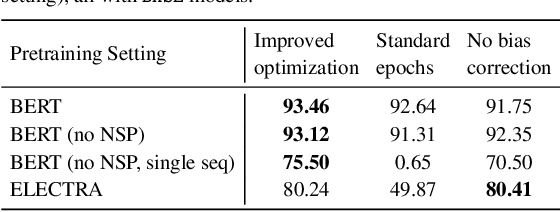
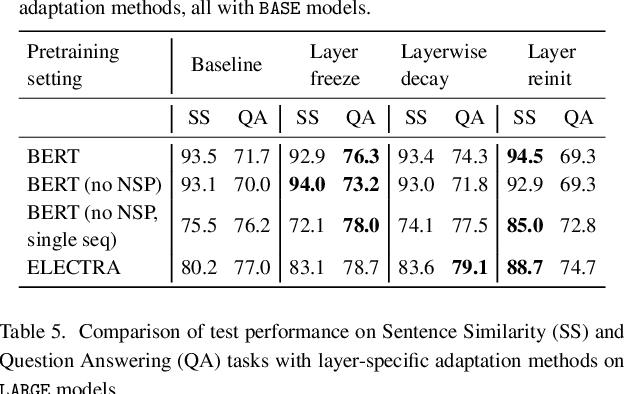
Abstract:Motivation: A perennial challenge for biomedical researchers and clinical practitioners is to stay abreast with the rapid growth of publications and medical notes. Natural language processing (NLP) has emerged as a promising direction for taming information overload. In particular, large neural language models facilitate transfer learning by pretraining on unlabeled text, as exemplified by the successes of BERT models in various NLP applications. However, fine-tuning such models for an end task remains challenging, especially with small labeled datasets, which are common in biomedical NLP. Results: We conduct a systematic study on fine-tuning stability in biomedical NLP. We show that finetuning performance may be sensitive to pretraining settings, especially in low-resource domains. Large models have potential to attain better performance, but increasing model size also exacerbates finetuning instability. We thus conduct a comprehensive exploration of techniques for addressing fine-tuning instability. We show that these techniques can substantially improve fine-tuning performance for lowresource biomedical NLP applications. Specifically, freezing lower layers is helpful for standard BERT-BASE models, while layerwise decay is more effective for BERT-LARGE and ELECTRA models. For low-resource text similarity tasks such as BIOSSES, reinitializing the top layer is the optimal strategy. Overall, domainspecific vocabulary and pretraining facilitate more robust models for fine-tuning. Based on these findings, we establish new state of the art on a wide range of biomedical NLP applications. Availability and implementation: To facilitate progress in biomedical NLP, we release our state-of-the-art pretrained and fine-tuned models: https://aka.ms/BLURB.
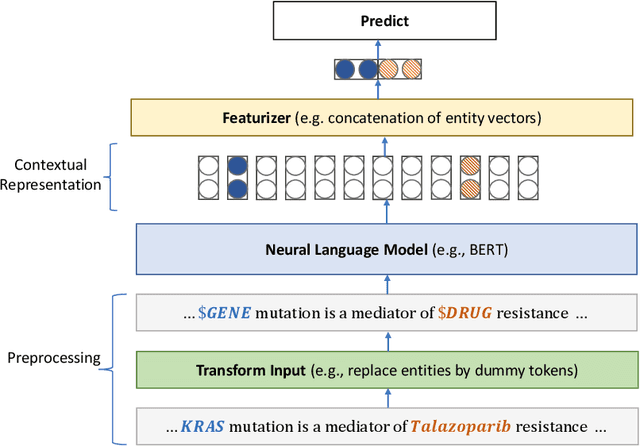
Modular Self-Supervision for Document-Level Relation Extraction
Sep 11, 2021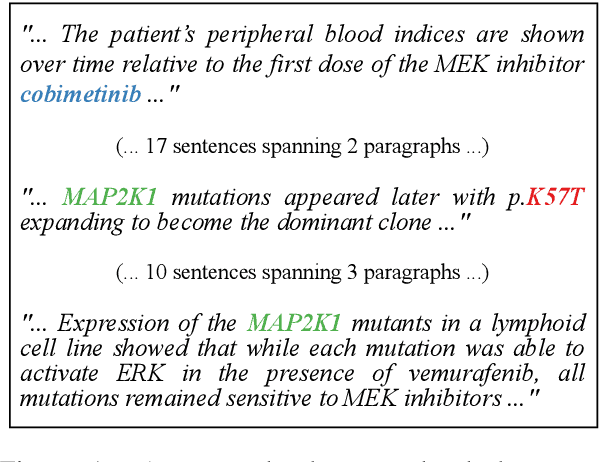
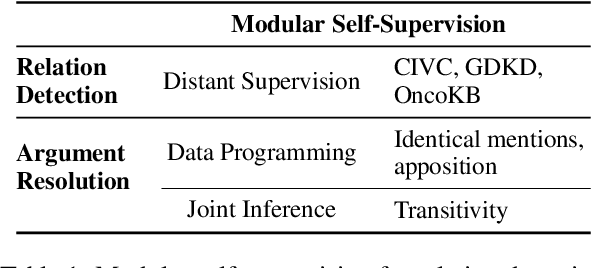
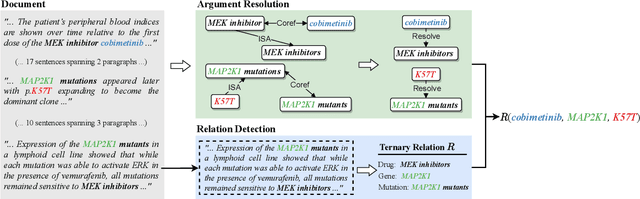
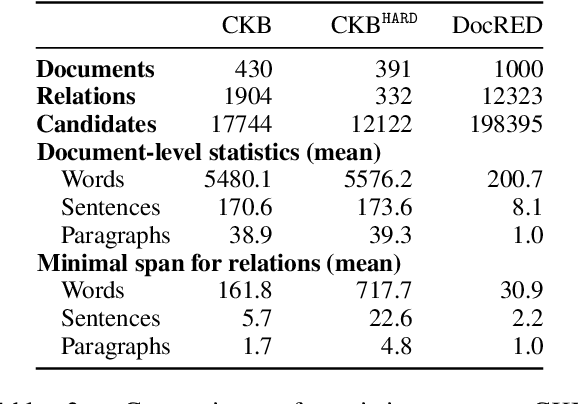
Abstract:Extracting relations across large text spans has been relatively underexplored in NLP, but it is particularly important for high-value domains such as biomedicine, where obtaining high recall of the latest findings is crucial for practical applications. Compared to conventional information extraction confined to short text spans, document-level relation extraction faces additional challenges in both inference and learning. Given longer text spans, state-of-the-art neural architectures are less effective and task-specific self-supervision such as distant supervision becomes very noisy. In this paper, we propose decomposing document-level relation extraction into relation detection and argument resolution, taking inspiration from Davidsonian semantics. This enables us to incorporate explicit discourse modeling and leverage modular self-supervision for each sub-problem, which is less noise-prone and can be further refined end-to-end via variational EM. We conduct a thorough evaluation in biomedical machine reading for precision oncology, where cross-paragraph relation mentions are prevalent. Our method outperforms prior state of the art, such as multi-scale learning and graph neural networks, by over 20 absolute F1 points. The gain is particularly pronounced among the most challenging relation instances whose arguments never co-occur in a paragraph.
Domain-Specific Pretraining for Vertical Search: Case Study on Biomedical Literature
Jun 25, 2021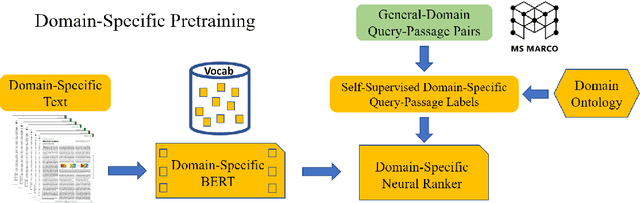
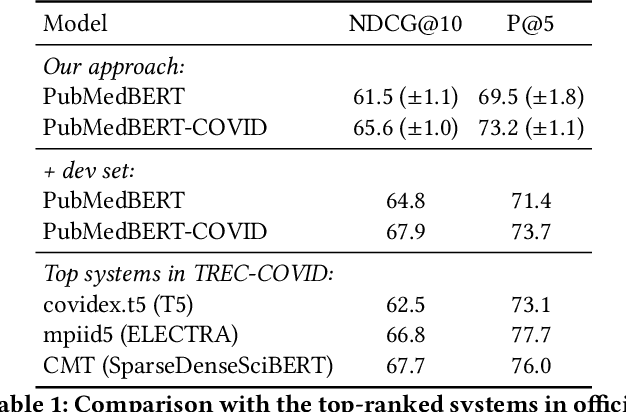
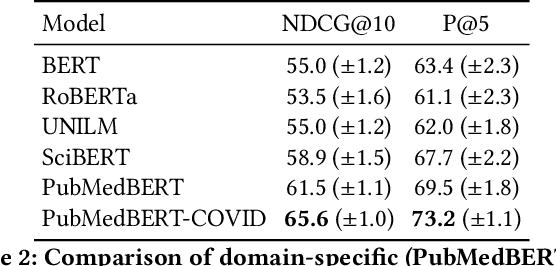
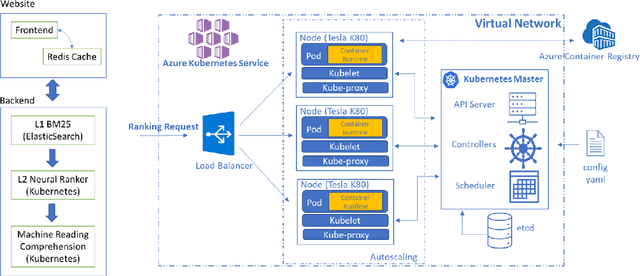
Abstract:Information overload is a prevalent challenge in many high-value domains. A prominent case in point is the explosion of the biomedical literature on COVID-19, which swelled to hundreds of thousands of papers in a matter of months. In general, biomedical literature expands by two papers every minute, totalling over a million new papers every year. Search in the biomedical realm, and many other vertical domains is challenging due to the scarcity of direct supervision from click logs. Self-supervised learning has emerged as a promising direction to overcome the annotation bottleneck. We propose a general approach for vertical search based on domain-specific pretraining and present a case study for the biomedical domain. Despite being substantially simpler and not using any relevance labels for training or development, our method performs comparably or better than the best systems in the official TREC-COVID evaluation, a COVID-related biomedical search competition. Using distributed computing in modern cloud infrastructure, our system can scale to tens of millions of articles on PubMed and has been deployed as Microsoft Biomedical Search, a new search experience for biomedical literature: https://aka.ms/biomedsearch.
Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing
Aug 20, 2020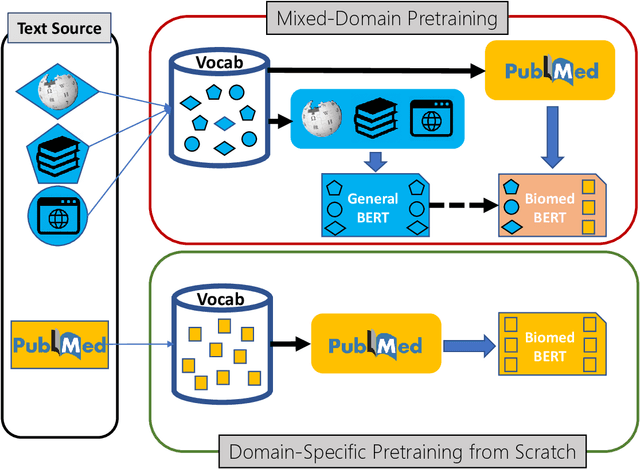
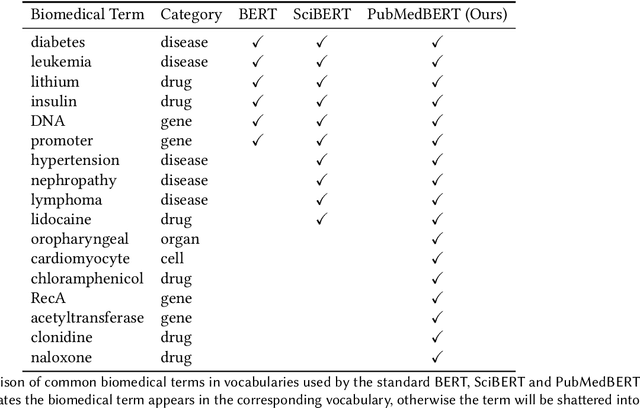
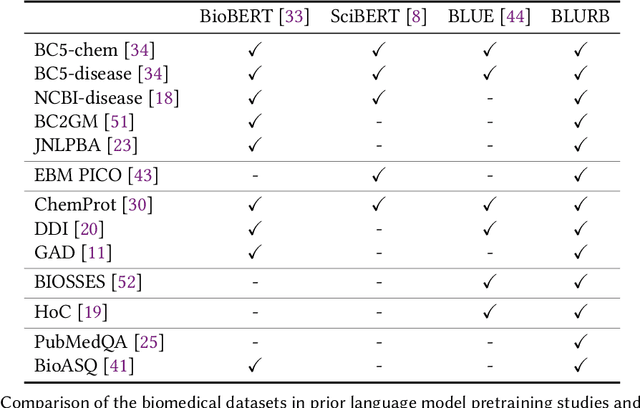
Abstract:Pretraining large neural language models, such as BERT, has led to impressive gains on many natural language processing (NLP) tasks. However, most pretraining efforts focus on general domain corpora, such as newswire and Web. A prevailing assumption is that even domain-specific pretraining can benefit by starting from general-domain language models. In this paper, we challenge this assumption by showing that for domains with abundant unlabeled text, such as biomedicine, pretraining language models from scratch results in substantial gains over continual pretraining of general-domain language models. To facilitate this investigation, we compile a comprehensive biomedical NLP benchmark from publicly-available datasets. Our experiments show that domain-specific pretraining serves as a solid foundation for a wide range of biomedical NLP tasks, leading to new state-of-the-art results across the board. Further, in conducting a thorough evaluation of modeling choices, both for pretraining and task-specific fine-tuning, we discover that some common practices are unnecessary with BERT models, such as using complex tagging schemes in named entity recognition (NER). To help accelerate research in biomedical NLP, we have released our state-of-the-art pretrained and task-specific models for the community, and created a leaderboard featuring our BLURB benchmark (short for Biomedical Language Understanding & Reasoning Benchmark) at https://aka.ms/BLURB.
ML4H Abstract Track 2019
Feb 05, 2020Abstract:A collection of the accepted abstracts for the Machine Learning for Health (ML4H) workshop at NeurIPS 2019. This index is not complete, as some accepted abstracts chose to opt-out of inclusion.
Cross-Language Aphasia Detection using Optimal Transport Domain Adaptation
Dec 04, 2019
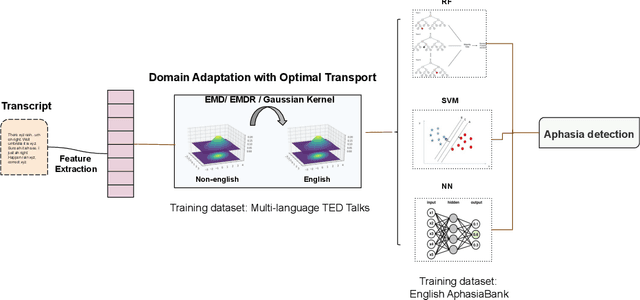
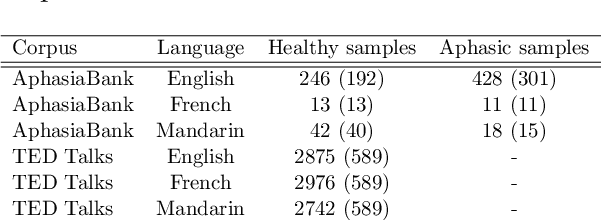
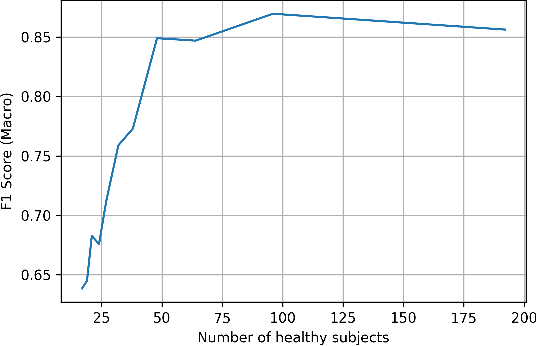
Abstract:Multi-language speech datasets are scarce and often have small sample sizes in the medical domain. Robust transfer of linguistic features across languages could improve rates of early diagnosis and therapy for speakers of low-resource languages when detecting health conditions from speech. We utilize out-of-domain, unpaired, single-speaker, healthy speech data for training multiple Optimal Transport (OT) domain adaptation systems. We learn mappings from other languages to English and detect aphasia from linguistic characteristics of speech, and show that OT domain adaptation improves aphasia detection over unilingual baselines for French (6% increased F1) and Mandarin (5% increased F1). Further, we show that adding aphasic data to the domain adaptation system significantly increases performance for both French and Mandarin, increasing the F1 scores further (10% and 8% increase in F1 scores for French and Mandarin, respectively, over unilingual baselines).
Feature Robustness in Non-stationary Health Records: Caveats to Deployable Model Performance in Common Clinical Machine Learning Tasks
Aug 02, 2019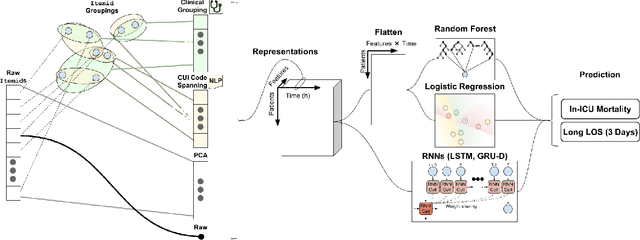
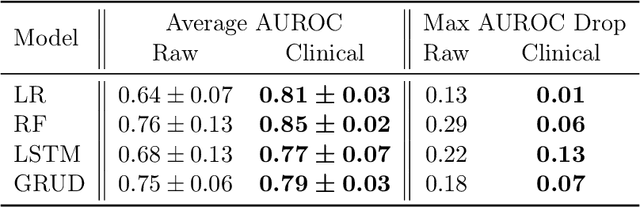
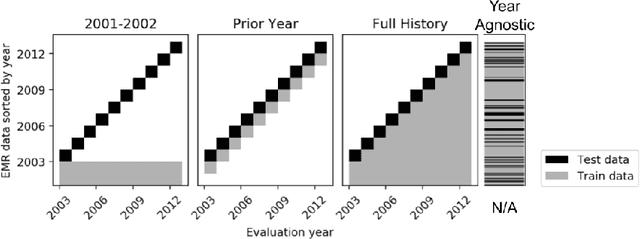
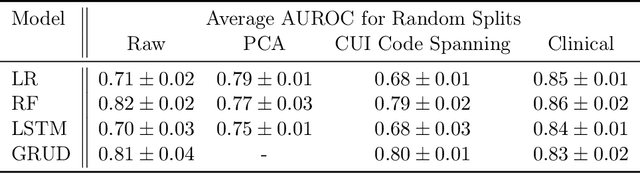
Abstract:When training clinical prediction models from electronic health records (EHRs), a key concern should be a model's ability to sustain performance over time when deployed, even as care practices, database systems, and population demographics evolve. Due to de-identification requirements, however, current experimental practices for public EHR benchmarks (such as the MIMIC-III critical care dataset) are time agnostic, assigning care records to train or test sets without regard for the actual dates of care. As a result, current benchmarks cannot assess how well models trained on one year generalise to another. In this work, we obtain a Limited Data Use Agreement to access year of care for each record in MIMIC and show that all tested state-of-the-art models decay in prediction quality when trained on historical data and tested on future data, particularly in response to a system-wide record-keeping change in 2008 (0.29 drop in AUROC for mortality prediction, 0.10 drop in AUROC for length-of-stay prediction with a random forest classifier). We further develop a simple yet effective mitigation strategy: by aggregating raw features into expert-defined clinical concepts, we see only a 0.06 drop in AUROC for mortality prediction and a 0.03 drop in AUROC for length-of-stay prediction. We demonstrate that this aggregation strategy outperforms other automatic feature preprocessing techniques aimed at increasing robustness to data drift. We release our aggregated representations and code to encourage more deployable clinical prediction models.
MIMIC-Extract: A Data Extraction, Preprocessing, and Representation Pipeline for MIMIC-III
Jul 19, 2019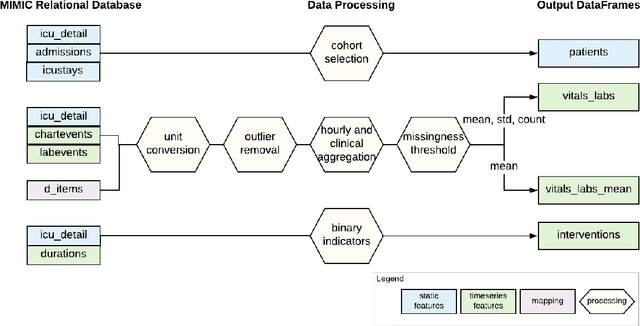
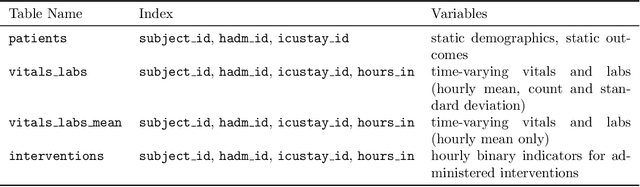
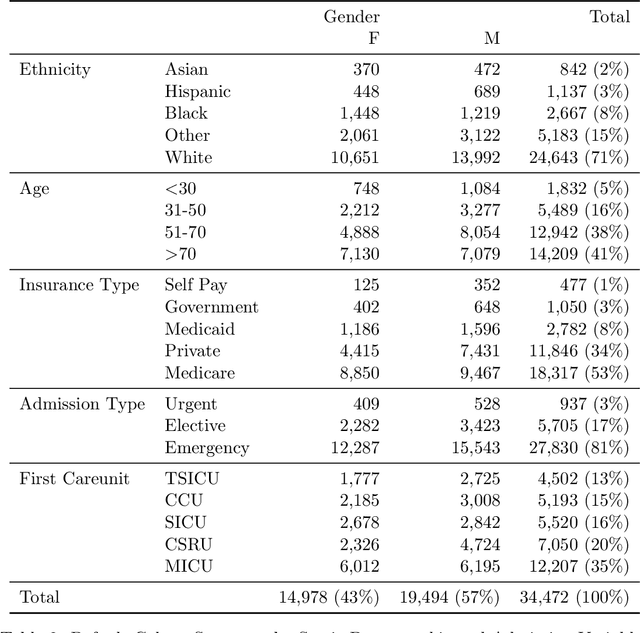
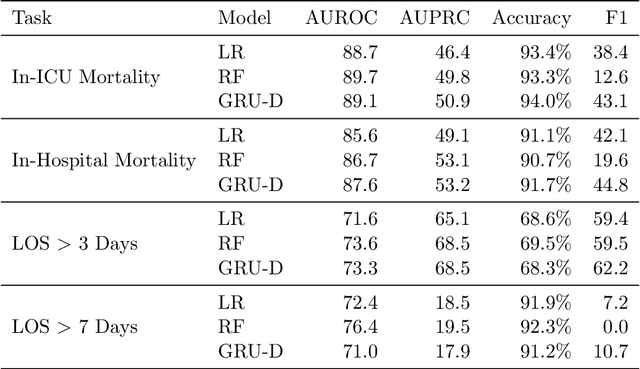
Abstract:Robust machine learning relies on access to data that can be used with standardized frameworks in important tasks and the ability to develop models whose performance can be reasonably reproduced. In machine learning for healthcare, the community faces reproducibility challenges due to a lack of publicly accessible data and a lack of standardized data processing frameworks. We present MIMIC-Extract, an open-source pipeline for transforming raw electronic health record (EHR) data for critical care patients contained in the publicly-available MIMIC-III database into dataframes that are directly usable in common machine learning pipelines. MIMIC-Extract addresses three primary challenges in making complex health records data accessible to the broader machine learning community. First, it provides standardized data processing functions, including unit conversion, outlier detection, and aggregating semantically equivalent features, thus accounting for duplication and reducing missingness. Second, it preserves the time series nature of clinical data and can be easily integrated into clinically actionable prediction tasks in machine learning for health. Finally, it is highly extensible so that other researchers with related questions can easily use the same pipeline. We demonstrate the utility of this pipeline by showcasing several benchmark tasks and baseline results.
Publicly Available Clinical BERT Embeddings
Apr 29, 2019
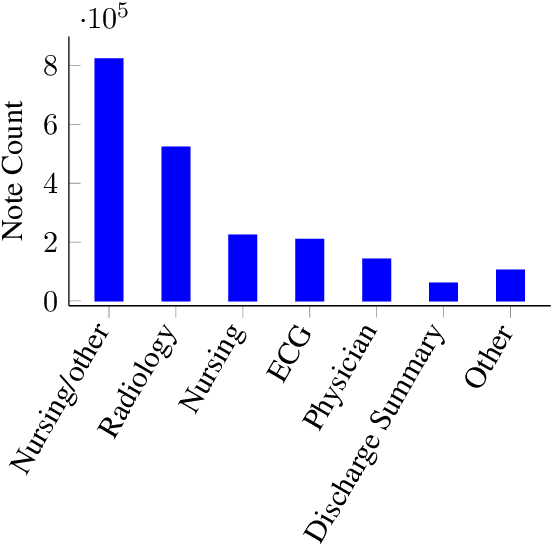
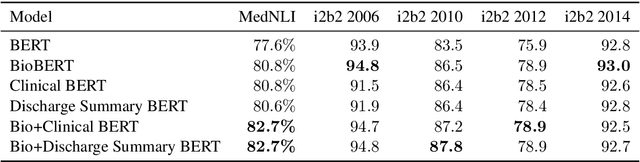
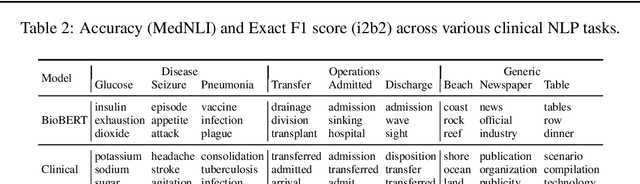
Abstract:Contextual word embedding models such as ELMo (Peters et al., 2018) and BERT (Devlin et al., 2018) have dramatically improved performance for many natural language processing (NLP) tasks in recent months. However, these models have been minimally explored on specialty corpora, such as clinical text; moreover, in the clinical domain, no publicly-available pre-trained BERT models yet exist. In this work, we address this need by exploring and releasing BERT models for clinical text: one for generic clinical text and another for discharge summaries specifically. We demonstrate that using a domain-specific model yields performance improvements on three common clinical NLP tasks as compared to nonspecific embeddings. These domain-specific models are not as performant on two clinical de-identification tasks, and argue that this is a natural consequence of the differences between de-identified source text and synthetically non de-identified task text.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge