Tianqi Yu
XDen-1K: A Density Field Dataset of Real-World Objects
Dec 11, 2025Abstract:A deep understanding of the physical world is a central goal for embodied AI and realistic simulation. While current models excel at capturing an object's surface geometry and appearance, they largely neglect its internal physical properties. This omission is critical, as properties like volumetric density are fundamental for predicting an object's center of mass, stability, and interaction dynamics in applications ranging from robotic manipulation to physical simulation. The primary bottleneck has been the absence of large-scale, real-world data. To bridge this gap, we introduce XDen-1K, the first large-scale, multi-modal dataset designed for real-world physical property estimation, with a particular focus on volumetric density. The core of this dataset consists of 1,000 real-world objects across 148 categories, for which we provide comprehensive multi-modal data, including a high-resolution 3D geometric model with part-level annotations and a corresponding set of real-world biplanar X-ray scans. Building upon this data, we introduce a novel optimization framework that recovers a high-fidelity volumetric density field of each object from its sparse X-ray views. To demonstrate its practical value, we add X-ray images as a conditioning signal to an existing segmentation network and perform volumetric segmentation. Furthermore, we conduct experiments on downstream robotics tasks. The results show that leveraging the dataset can effectively improve the accuracy of center-of-mass estimation and the success rate of robotic manipulation. We believe XDen-1K will serve as a foundational resource and a challenging new benchmark, catalyzing future research in physically grounded visual inference and embodied AI.
CMG-Net: Robust Normal Estimation for Point Clouds via Chamfer Normal Distance and Multi-scale Geometry
Dec 14, 2023



Abstract:This work presents an accurate and robust method for estimating normals from point clouds. In contrast to predecessor approaches that minimize the deviations between the annotated and the predicted normals directly, leading to direction inconsistency, we first propose a new metric termed Chamfer Normal Distance to address this issue. This not only mitigates the challenge but also facilitates network training and substantially enhances the network robustness against noise. Subsequently, we devise an innovative architecture that encompasses Multi-scale Local Feature Aggregation and Hierarchical Geometric Information Fusion. This design empowers the network to capture intricate geometric details more effectively and alleviate the ambiguity in scale selection. Extensive experiments demonstrate that our method achieves the state-of-the-art performance on both synthetic and real-world datasets, particularly in scenarios contaminated by noise. Our implementation is available at https://github.com/YingruiWoo/CMG-Net_Pytorch.
DeepACC:Automate Chromosome Classification based on Metaphase Images using Deep Learning Framework Fused with Prior Knowledge
Jun 28, 2020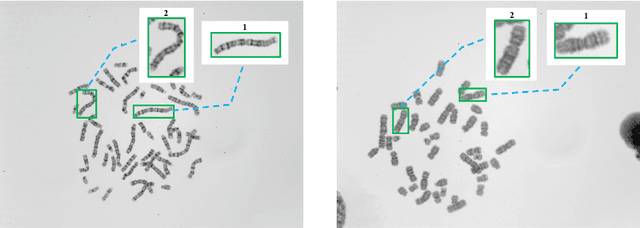
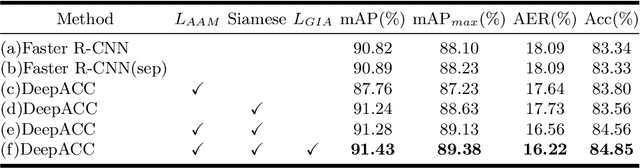
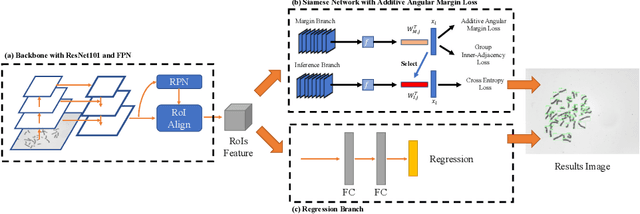

Abstract:Chromosome classification is an important but difficult and tedious task in karyotyping. Previous methods only classify manually segmented single chromosome, which is far from clinical practice. In this work, we propose a detection based method, DeepACC, to locate and fine classify chromosomes simultaneously based on the whole metaphase image. We firstly introduce the Additive Angular Margin Loss to enhance the discriminative power of model. To alleviate batch effects, we transform decision boundary of each class case-by-case through a siamese network which make full use of prior knowledges that chromosomes usually appear in pairs. Furthermore, we take the clinically seven group criterion as a prior knowledge and design an additional Group Inner-Adjacency Loss to further reduce inter-class similarities. 3390 metaphase images from clinical laboratory are collected and labelled to evaluate the performance. Results show that the new design brings encouraging performance gains comparing to the state-of-the-art baselines.
DeepACE: Automated Chromosome Enumeration in Metaphase Cell Images Using Deep Convolutional Neural Networks
Oct 12, 2019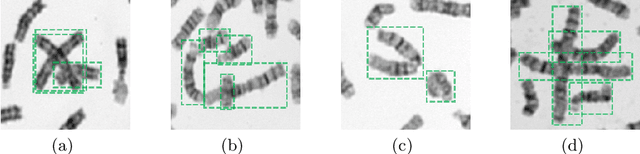
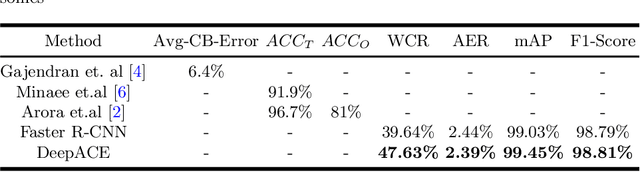
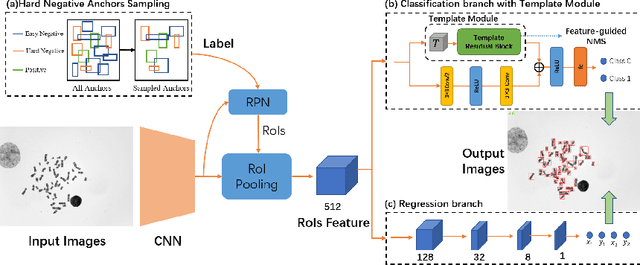
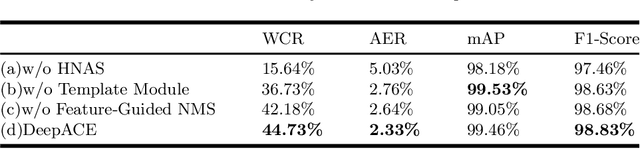
Abstract:Chromosome enumeration is an important but tedious procedure in karyotyping analysis. In this paper, to automate the enumeration process, we developed a chromosome enumeration framework, DeepACE, based on the region based object detection scheme. Firstly, the ability of region proposal network is enhanced by a newly proposed Hard Negative Anchors Sampling to extract unapparent but important information about highly confusing partial chromosomes. Next, to alleviate serious occlusion problems, we novelly introduced a weakly-supervised mechanism by adding a Template Module into classification branch to heuristically separate overlapped chromosomes. The template features are further incorporated into the NMS procedure to further improve the detection of overlapping chromosomes. In the newly collected clinical dataset, the proposed method outperform all the previous method, yielding an mAP with respect to chromosomes as 99.45, and the error rate is about 2.4%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge