Shadi Albarqouni
University Hospital Bonn, Venusberg-Campus 1, D-53127, Bonn, Germany, Helmholtz Munich, Ingolstädter Landstraße 1, D-85764, Neuherberg, Germany, Technical University of Munich, Boltzmannstr. 3, D-85748 Garching, Germany
6D Camera Relocalization in Ambiguous Scenes via Continuous Multimodal Inference
Apr 09, 2020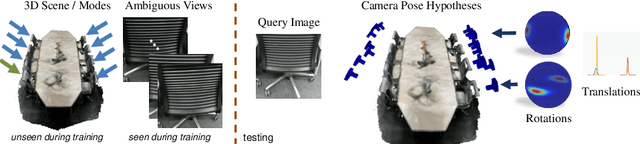
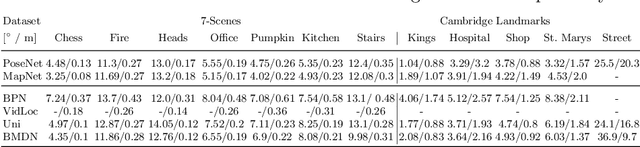
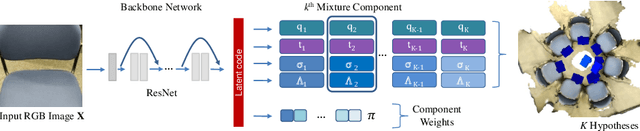
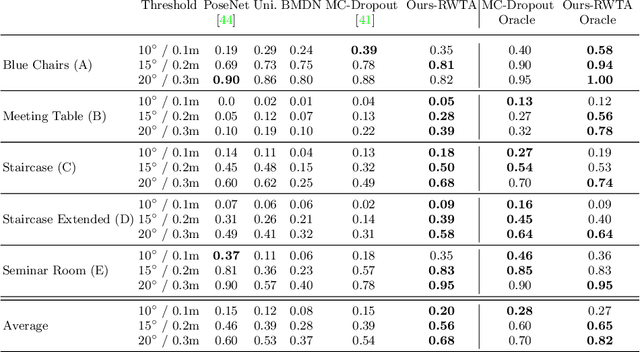
Abstract:We present a multimodal camera relocalization framework that captures ambiguities and uncertainties with continuous mixture models defined on the manifold of camera poses. In highly ambiguous environments, which can easily arise due to symmetries and repetitive structures in the scene, computing one plausible solution (what most state-of-the-art methods currently regress) may not be sufficient. Instead we predict multiple camera pose hypotheses as well as the respective uncertainty for each prediction. Towards this aim, we use Bingham distributions, to model the orientation of the camera pose, and a multivariate Gaussian to model the position, with an end-to-end deep neural network. By incorporating a Winner-Takes-All training scheme, we finally obtain a mixture model that is well suited for explaining ambiguities in the scene, yet does not suffer from mode collapse, a common problem with mixture density networks. We introduce a new dataset specifically designed to foster camera localization research in ambiguous environments and exhaustively evaluate our method on synthetic as well as real data on both ambiguous scenes and on non-ambiguous benchmark datasets. We plan to release our code and dataset under $\href{https://multimodal3dvision.github.io}{multimodal3dvision.github.io}$.
Autoencoders for Unsupervised Anomaly Segmentation in Brain MR Images: A Comparative Study
Apr 08, 2020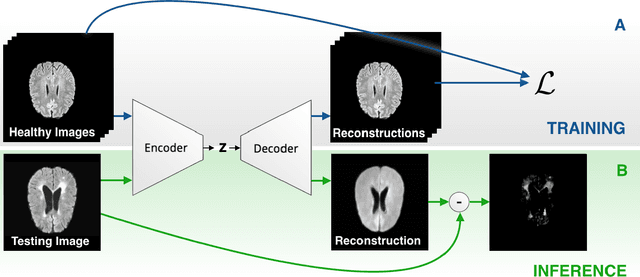
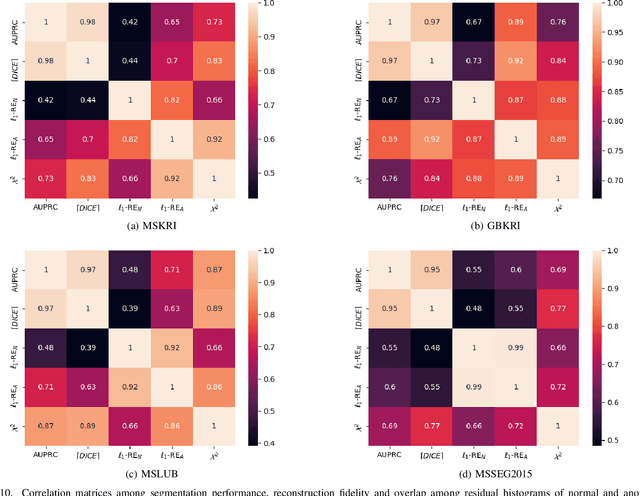
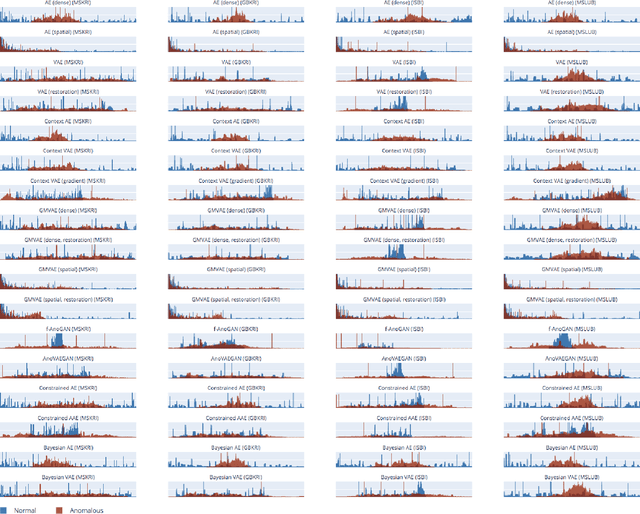
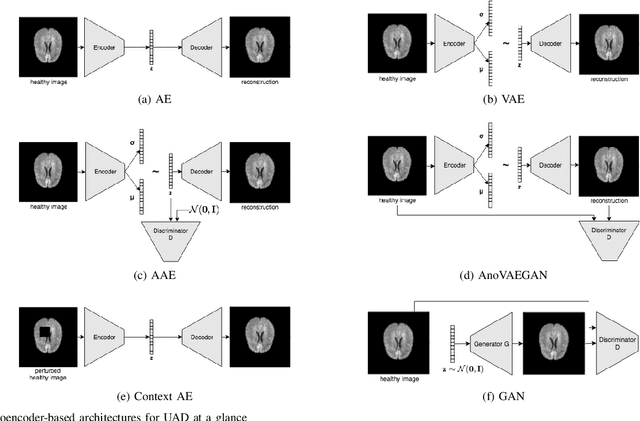
Abstract:Deep unsupervised representation learning has recently led to new approaches in the field of Unsupervised Anomaly Detection (UAD) in brain MRI. The main principle behind these works is to learn a model of normal anatomy by learning to compress and recover healthy data. This allows to spot abnormal structures from erroneous recoveries of compressed, potentially anomalous samples. The concept is of great interest to the medical image analysis community as it i) relieves from the need of vast amounts of manually segmented training data---a necessity for and pitfall of current supervised Deep Learning---and ii) theoretically allows to detect arbitrary, even rare pathologies which supervised approaches might fail to find. To date, the experimental design of most works hinders a valid comparison, because i) they are evaluated against different datasets and different pathologies, ii) use different image resolutions and iii) different model architectures with varying complexity. The intent of this work is to establish comparability among recent methods by utilizing a single architecture, a single resolution and the same dataset(s). Besides providing a ranking of the methods, we also try to answer questions like i) how many healthy training subjects are needed to model normality and ii) if the reviewed approaches are also sensitive to domain shift. Further, we identify open challenges and provide suggestions for future community efforts and research directions.
Fairness by Learning Orthogonal Disentangled Representations
Mar 22, 2020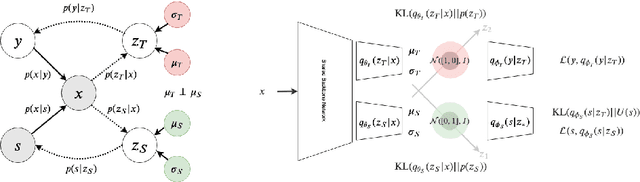
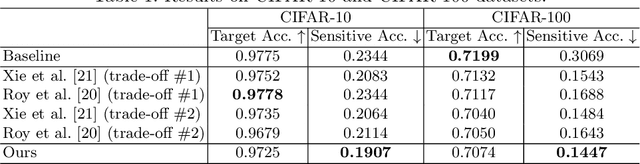
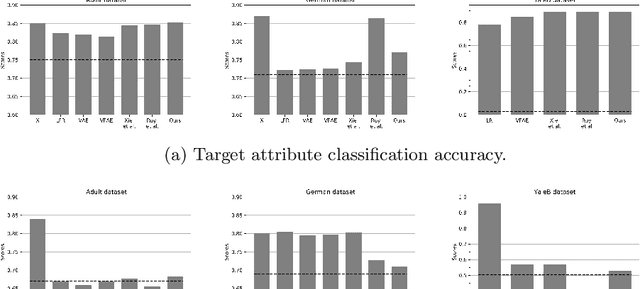
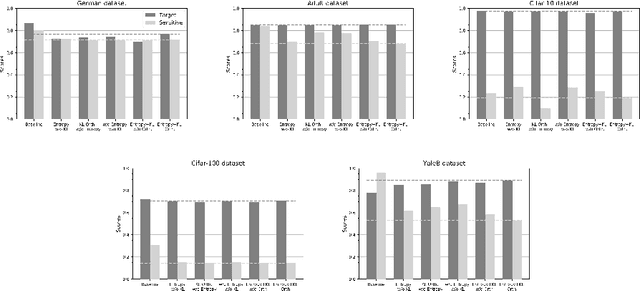
Abstract:Learning discriminative powerful representations is a crucial step for machine learning systems. Introducing invariance against arbitrary nuisance or sensitive attributes while performing well on specific tasks is an important problem in representation learning. This is mostly approached by purging the sensitive information from learned representations. In this paper, we propose a novel disentanglement approach to invariant representation problem. We disentangle the meaningful and sensitive representations by enforcing orthogonality constraints as a proxy for independence. We explicitly enforce the meaningful representation to be agnostic to sensitive information by entropy maximization. The proposed approach is evaluated on five publicly available datasets and compared with state of the art methods for learning fairness and invariance achieving the state of the art performance on three datasets and comparable performance on the rest. Further, we perform an ablative study to evaluate the effect of each component.
ROAM: Random Layer Mixup for Semi-Supervised Learning in Medical Imaging
Mar 20, 2020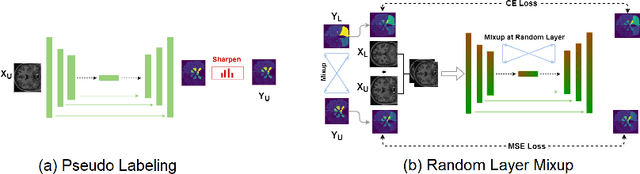
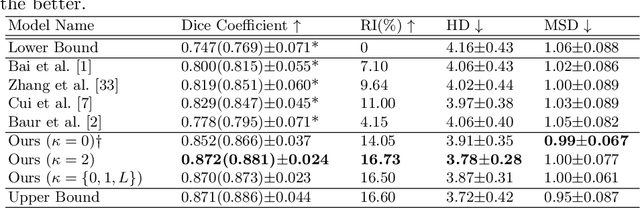
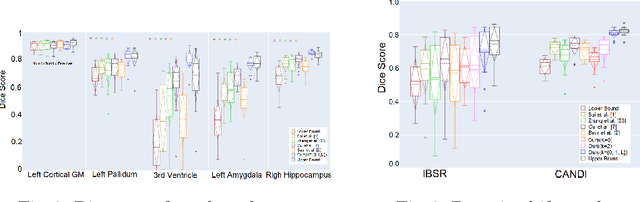
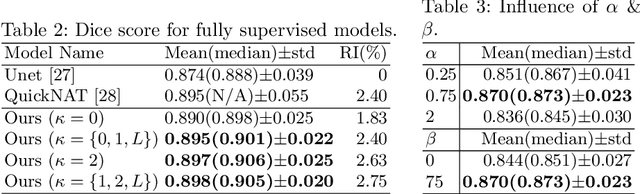
Abstract:Medical image segmentation is one of the major challenges addressed by machine learning methods. Yet, deep learning methods profoundly depend on a huge amount of annotated data which is time-consuming and costly. Though semi-supervised learning methods approach this problem by leveraging an abundant amount of unlabeled data along with a small amount of labeled data in the training process. Recently, MixUp regularizer [32] has been successfully introduced to semi-supervised learning methods showing superior performance [3]. MixUp augments the model with new data points through linear interpolation of the data at the input space. In this paper, we argue that this option is limited, instead, we propose ROAM, a random layer mixup, which encourages the network to be less confident for interpolated data points at randomly selected space. Hence, avoids over-fitting and enhances the generalization ability. We validate our method on publicly available datasets on whole-brain image segmentation (MALC) achieving state-of-the-art results in fully supervised (89.8%) and semi-supervised (87.2%) settings with relative improvement up to 2.75% and 16.73%, respectively.
The Future of Digital Health with Federated Learning
Mar 18, 2020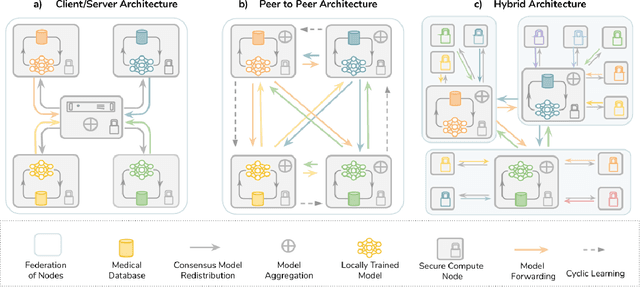
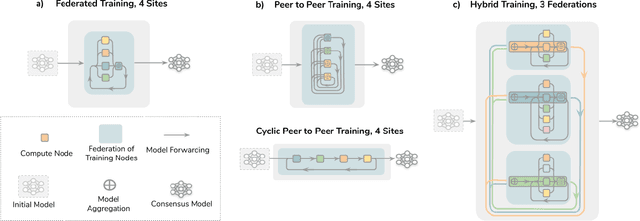
Abstract:Data-driven Machine Learning has emerged as a promising approach for building accurate and robust statistical models from medical data, which is collected in huge volumes by modern healthcare systems. Existing medical data is not fully exploited by ML primarily because it sits in data silos and privacy concerns restrict access to this data. However, without access to sufficient data, ML will be prevented from reaching its full potential and, ultimately, from making the transition from research to clinical practice. This paper considers key factors contributing to this issue, explores how Federated Learning (FL) may provide a solution for the future of digital health and highlights the challenges and considerations that need to be addressed.
A learning without forgetting approach to incorporate artifact knowledge in polyp localization tasks
Feb 11, 2020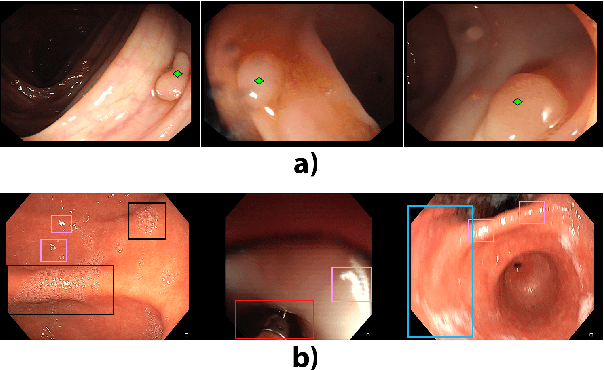
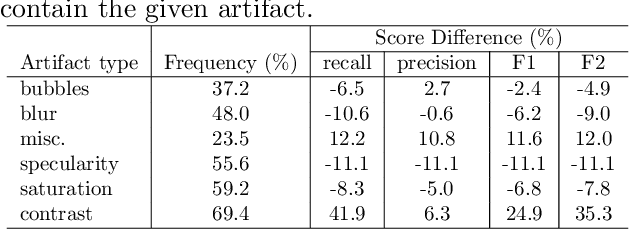
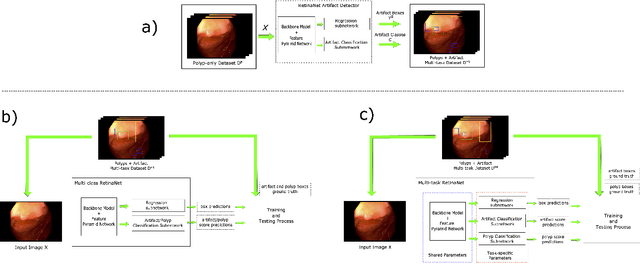
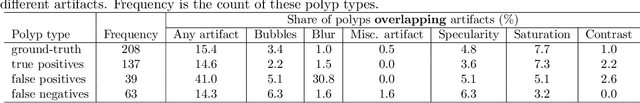
Abstract:Colorectal polyps are abnormalities in the colon tissue that can develop into colorectal cancer. The survival rate for patients is higher when the disease is detected at an early stage and polyps can be removed before they develop into malignant tumors. Deep learning methods have become the state of art in automatic polyp detection. However, the performance of current models heavily relies on the size and quality of the training datasets. Endoscopic video sequences tend to be corrupted by different artifacts affecting visibility and hence, the detection rates. In this work, we analyze the effects that artifacts have in the polyp localization problem. For this, we evaluate the RetinaNet architecture, originally defined for object localization. We also define a model inspired by the learning without forgetting framework, which allows us to employ artifact detection knowledge in the polyp localization problem. Finally, we perform several experiments to analyze the influence of the artifacts in the performance of these models. To our best knowledge, this is the first extensive analysis of the influence of artifact in polyp localization and the first work incorporating learning without forgetting ideas for simultaneous artifact and polyp localization tasks.
Learn to Estimate Labels Uncertainty for Quality Assurance
Sep 17, 2019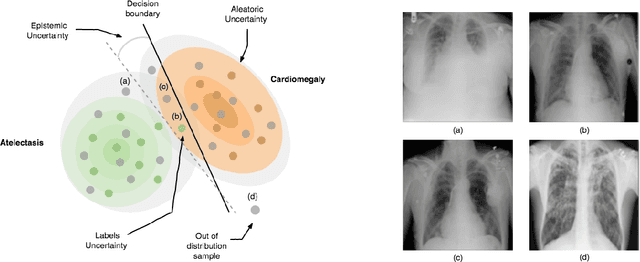
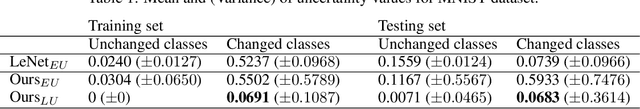

Abstract:Deep Learning sets the state-of-the-art in many challenging tasks showing outstanding performance in a broad range of applications. Despite its success, it still lacks robustness hindering its adoption in medical applications. Modeling uncertainty, through Bayesian Inference and Monte-Carlo dropout, has been successfully introduced for better understanding the underlying deep learning models. Yet, another important source of uncertainty, coming from the inter-observer variability, has not been thoroughly addressed in the literature. In this paper, we introduce labels uncertainty which better suits medical applications and show that modeling such uncertainty together with epistemic uncertainty is of high interest for quality control and referral systems.
Learn to Segment Organs with a Few Bounding Boxes
Sep 17, 2019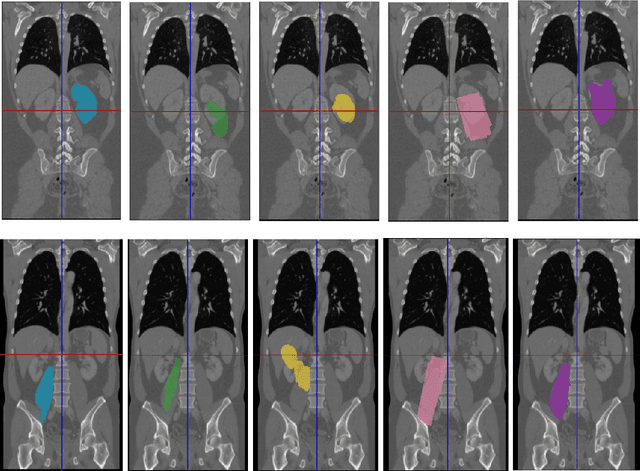
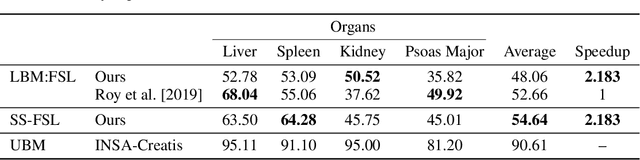
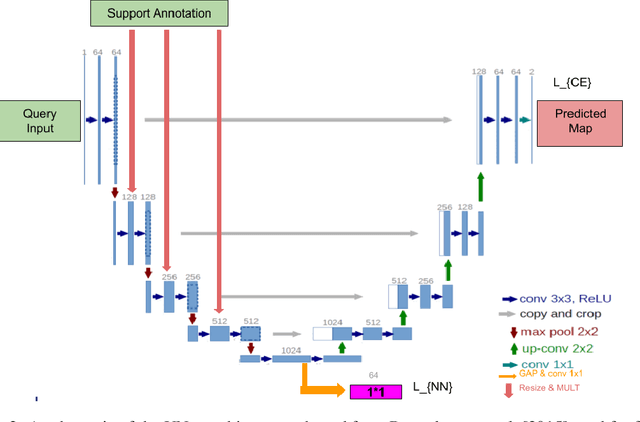
Abstract:Semantic segmentation is an import task in the medical field to identify the exact extent and orientation of significant structures like organs and pathology. Deep neural networks can perform this task well by leveraging the information from a large well-labeled data-set. This paper aims to present a method that mitigates the necessity of an extensive well-labeled data-set. This method also addresses semi-supervision by enabling segmentation based on bounding box annotations, avoiding the need for full pixel-level annotations. The network presented consists of a single U-Net based unbranched architecture that generates a few-shot segmentation for an unseen human organ using just 4 example annotations of that specific organ. The network is trained by alternately minimizing the nearest neighbor loss for prototype learning and a weighted cross-entropy loss for segmentation learning to perform a fast 3D segmentation with a median score of 54.64%.
Image to Images Translation for Multi-Task Organ Segmentation and Bone Suppression in Chest X-Ray Radiography
Jun 24, 2019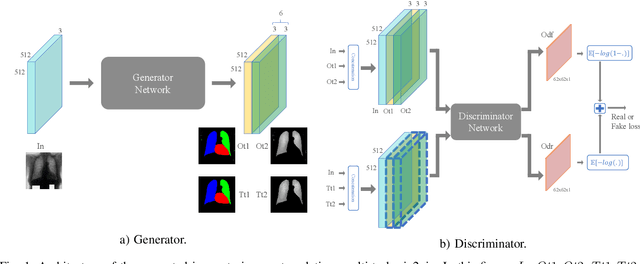
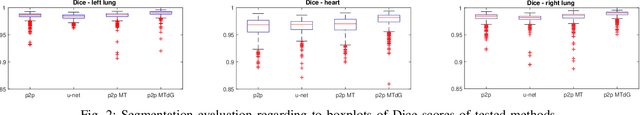
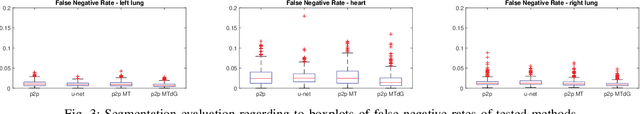
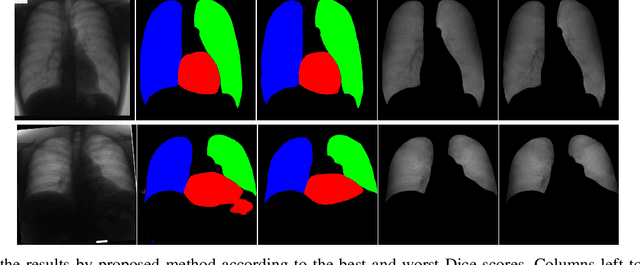
Abstract:Chest X-ray radiography is one of the earliest medical imaging technologies and remains one of the most widely-used for the diagnosis, screening and treatment follow up of diseases related to lungs and heart. The literature in this field of research reports many interesting studies dealing with the challenging tasks of bone suppression and organ segmentation but performed separately, limiting any learning that comes with the consolidation of parameters that could optimize both processes. Although image processing could facilitate computer aided diagnosis, machine learning seems more amenable in dealing with the many parameters one would have to contend with to yield an near optimal classification or decision-making process. This study, and for the first time, introduces a multitask deep learning model that generates simultaneously the bone-suppressed image and the organ segmented image, minimizing as a consequence the number of parameters the model has to deal with and optimizing the processing time as well; while at the same time exploiting the interplay in these parameters so as to benefit the performance of both tasks. The design architecture of this model, which relies on a conditional generative adversarial network, reveals the process on how we managed to modify the well-established pix2pix network to fit the need for multitasking and hence extending the standard image-to-image network to the new image-to-images architecture. Dilated convolutions are also used to improve the results through a more effective receptive field assessment. A comparison of the proposed approach to state-of-the-art algorithms is provided to gauge the merits of the proposed approach.
Perceptual Embedding Consistency for Seamless Reconstruction of Tilewise Style Transfer
Jun 03, 2019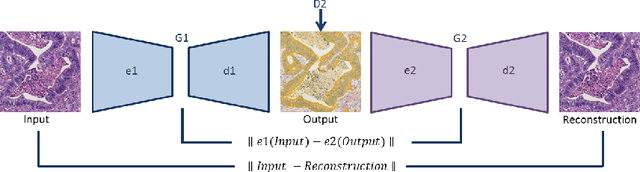

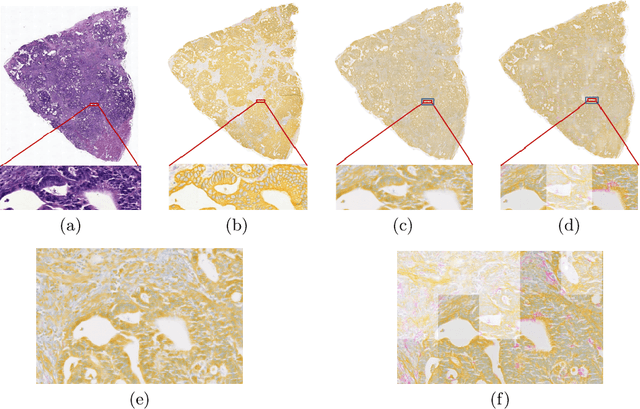
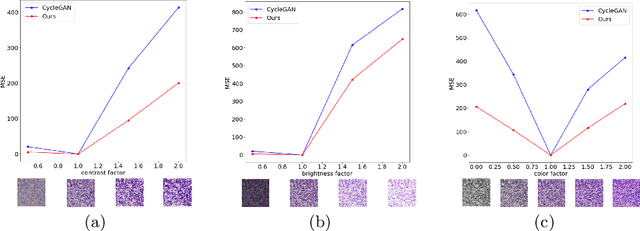
Abstract:Style transfer is a field with growing interest and use cases in deep learning. Recent work has shown Generative Adversarial Networks(GANs) can be used to create realistic images of virtually stained slide images in digital pathology with clinically validated interpretability. Digital pathology images are typically of extremely high resolution, making tilewise analysis necessary for deep learning applications. It has been shown that image generators with instance normalization can cause a tiling artifact when a large image is reconstructed from the tilewise analysis. We introduce a novel perceptual embedding consistency loss significantly reducing the tiling artifact created in the reconstructed whole slide image (WSI). We validate our results by comparing virtually stained slide images with consecutive real stained tissue slide images. We also demonstrate that our model is more robust to contrast, color and brightness perturbations by running comparative sensitivity analysis tests.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge