Mirabela Rusu
ConFUDA: Contrastive Fewshot Unsupervised Domain Adaptation for Medical Image Segmentation
Jun 08, 2022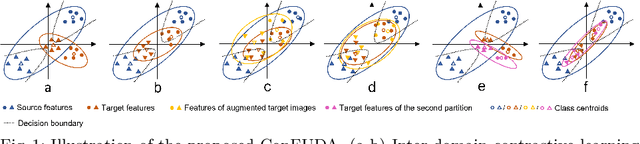
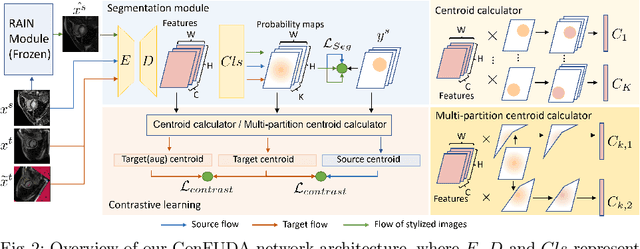
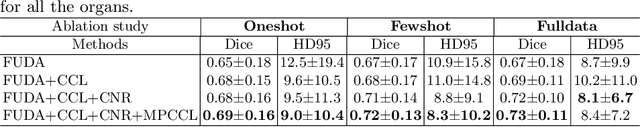
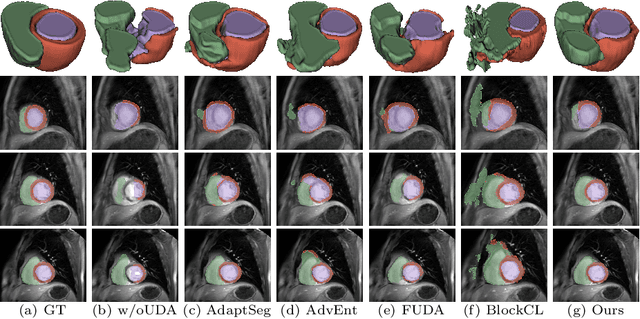
Abstract:Unsupervised domain adaptation (UDA) aims to transfer knowledge learned from a labeled source domain to an unlabeled target domain. Contrastive learning (CL) in the context of UDA can help to better separate classes in feature space. However, in image segmentation, the large memory footprint due to the computation of the pixel-wise contrastive loss makes it prohibitive to use. Furthermore, labeled target data is not easily available in medical imaging, and obtaining new samples is not economical. As a result, in this work, we tackle a more challenging UDA task when there are only a few (fewshot) or a single (oneshot) image available from the target domain. We apply a style transfer module to mitigate the scarcity of target samples. Then, to align the source and target features and tackle the memory issue of the traditional contrastive loss, we propose the centroid-based contrastive learning (CCL) and a centroid norm regularizer (CNR) to optimize the contrastive pairs in both direction and magnitude. In addition, we propose multi-partition centroid contrastive learning (MPCCL) to further reduce the variance in the target features. Fewshot evaluation on MS-CMRSeg dataset demonstrates that ConFUDA improves the segmentation performance by 0.34 of the Dice score on the target domain compared with the baseline, and 0.31 Dice score improvement in a more rigorous oneshot setting.
Image quality assessment for machine learning tasks using meta-reinforcement learning
Mar 27, 2022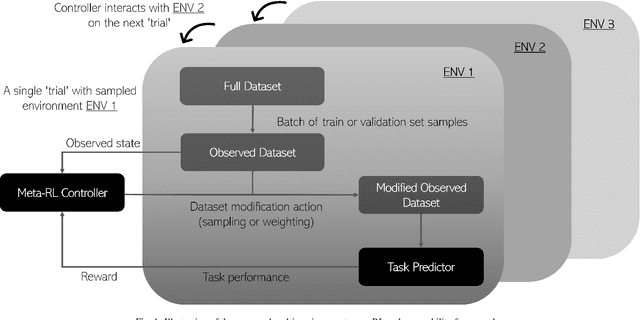
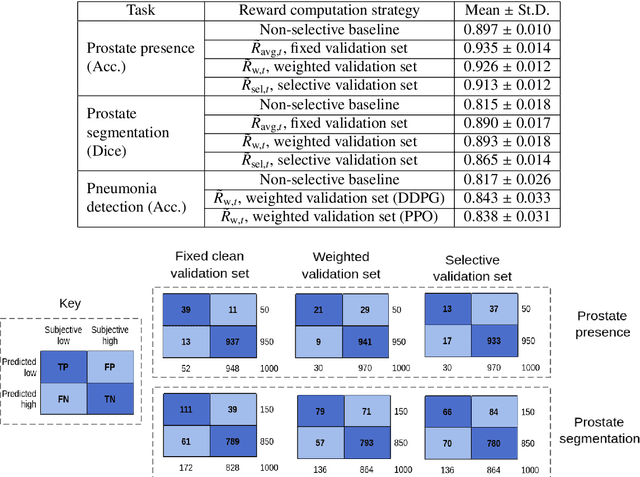
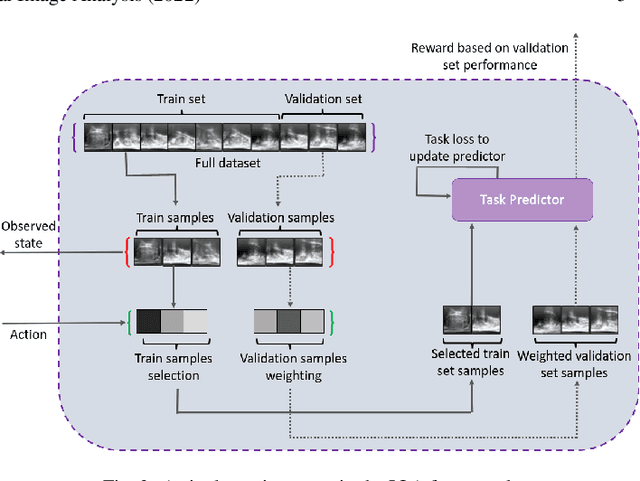
Abstract:In this paper, we consider image quality assessment (IQA) as a measure of how images are amenable with respect to a given downstream task, or task amenability. When the task is performed using machine learning algorithms, such as a neural-network-based task predictor for image classification or segmentation, the performance of the task predictor provides an objective estimate of task amenability. In this work, we use an IQA controller to predict the task amenability which, itself being parameterised by neural networks, can be trained simultaneously with the task predictor. We further develop a meta-reinforcement learning framework to improve the adaptability for both IQA controllers and task predictors, such that they can be fine-tuned efficiently on new datasets or meta-tasks. We demonstrate the efficacy of the proposed task-specific, adaptable IQA approach, using two clinical applications for ultrasound-guided prostate intervention and pneumonia detection on X-ray images.
* Accepted to Medical Image Analysis; Final published version available at: https://doi.org/10.1016/j.media.2022.102427
Image quality assessment by overlapping task-specific and task-agnostic measures: application to prostate multiparametric MR images for cancer segmentation
Feb 20, 2022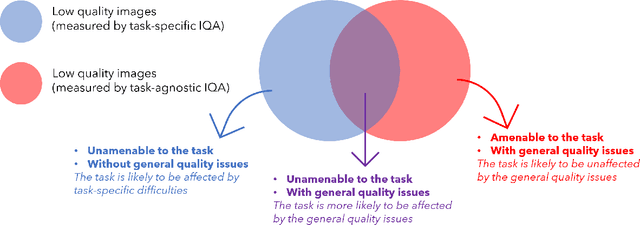
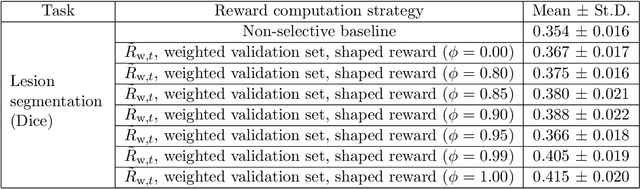
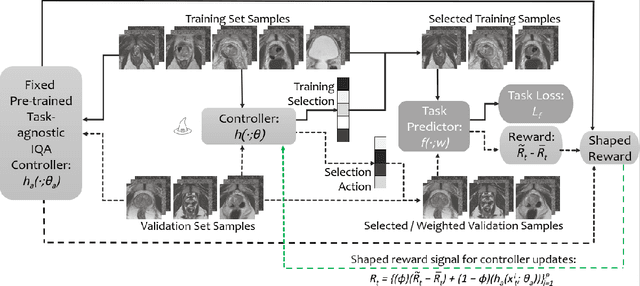
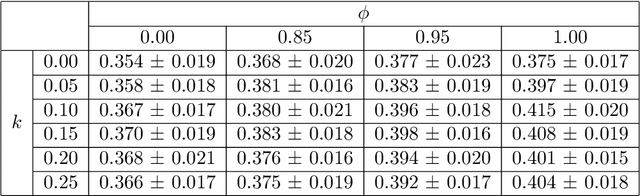
Abstract:Image quality assessment (IQA) in medical imaging can be used to ensure that downstream clinical tasks can be reliably performed. Quantifying the impact of an image on the specific target tasks, also named as task amenability, is needed. A task-specific IQA has recently been proposed to learn an image-amenability-predicting controller simultaneously with a target task predictor. This allows for the trained IQA controller to measure the impact an image has on the target task performance, when this task is performed using the predictor, e.g. segmentation and classification neural networks in modern clinical applications. In this work, we propose an extension to this task-specific IQA approach, by adding a task-agnostic IQA based on auto-encoding as the target task. Analysing the intersection between low-quality images, deemed by both the task-specific and task-agnostic IQA, may help to differentiate the underpinning factors that caused the poor target task performance. For example, common imaging artefacts may not adversely affect the target task, which would lead to a low task-agnostic quality and a high task-specific quality, whilst individual cases considered clinically challenging, which can not be improved by better imaging equipment or protocols, is likely to result in a high task-agnostic quality but a low task-specific quality. We first describe a flexible reward shaping strategy which allows for the adjustment of weighting between task-agnostic and task-specific quality scoring. Furthermore, we evaluate the proposed algorithm using a clinically challenging target task of prostate tumour segmentation on multiparametric magnetic resonance (mpMR) images, from 850 patients. The proposed reward shaping strategy, with appropriately weighted task-specific and task-agnostic qualities, successfully identified samples that need re-acquisition due to defected imaging process.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021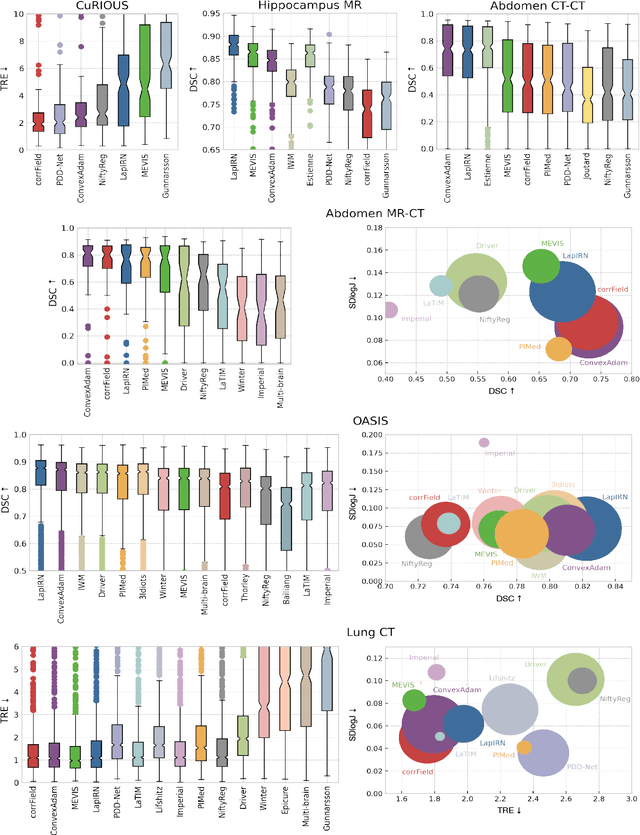
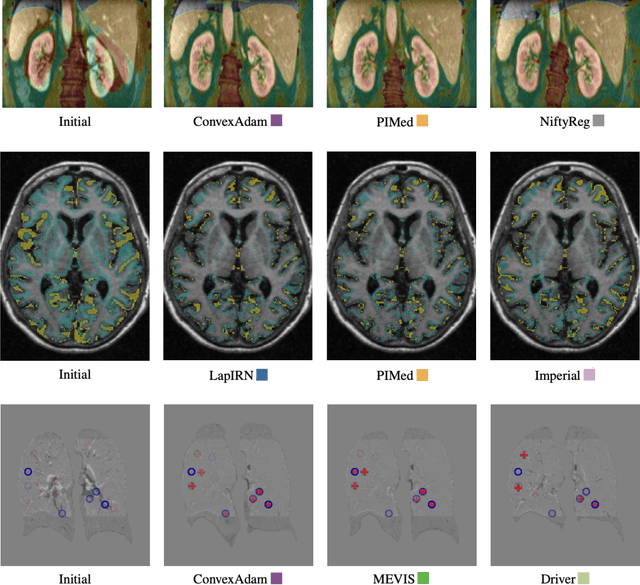
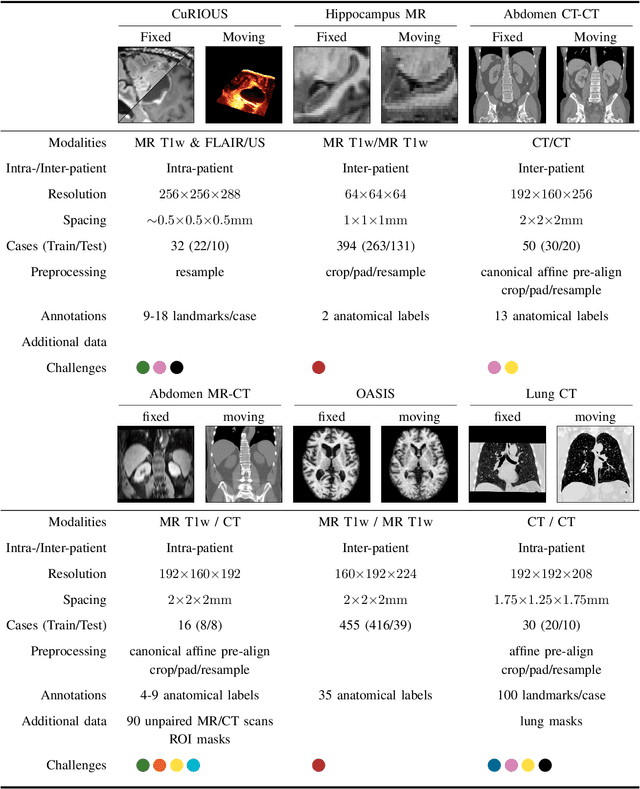
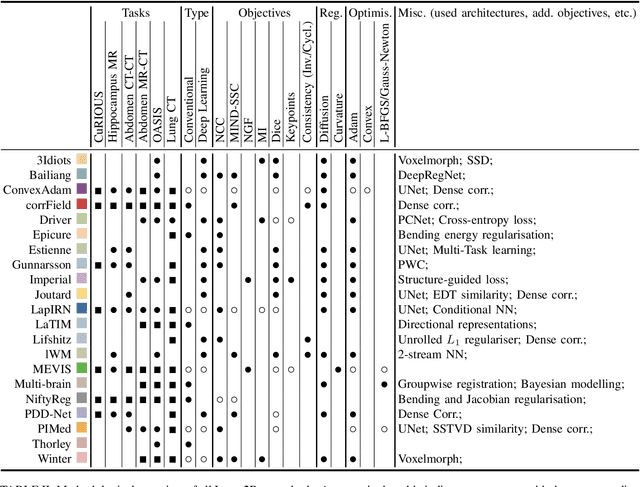
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Bridging the gap between prostate radiology and pathology through machine learning
Dec 03, 2021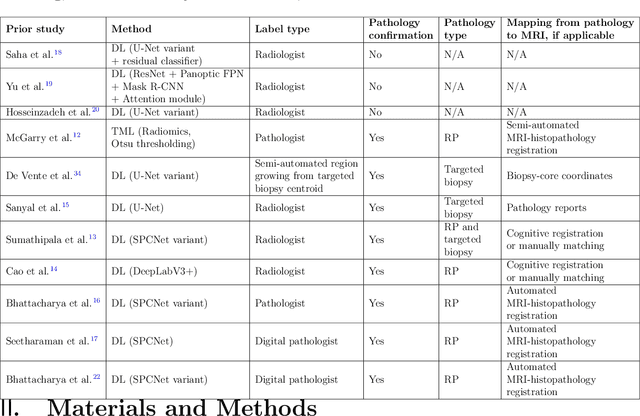
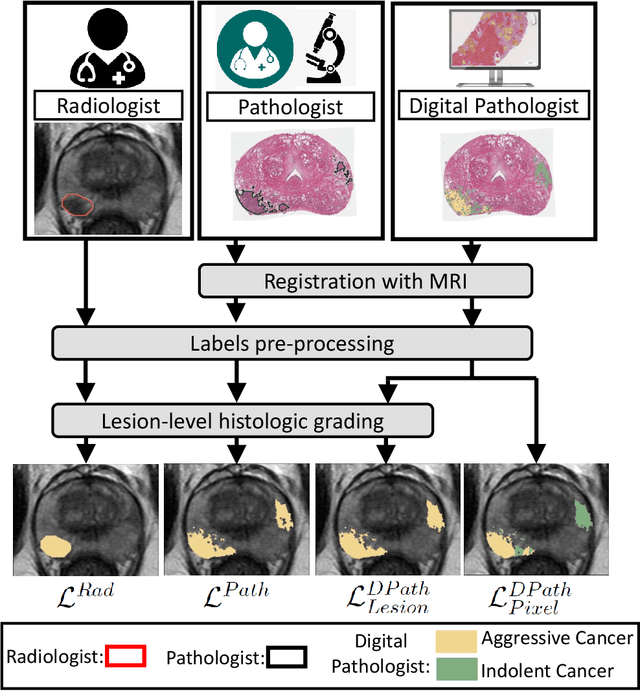
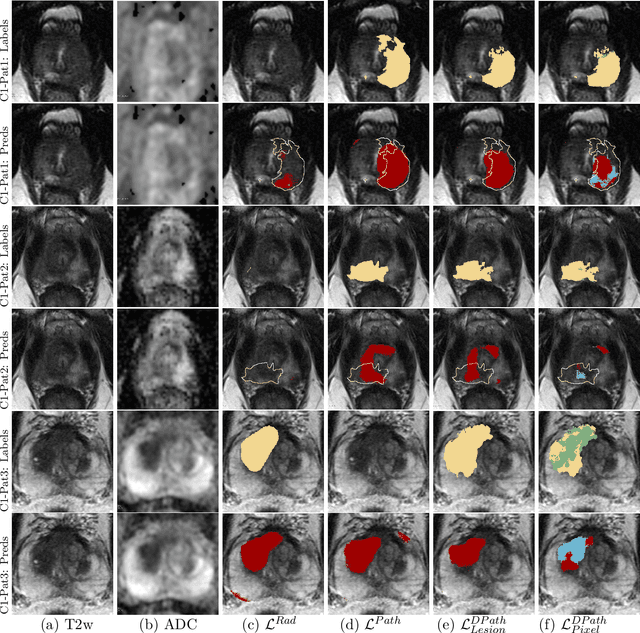
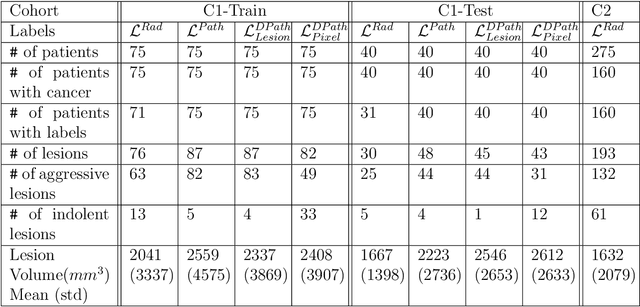
Abstract:Prostate cancer is the second deadliest cancer for American men. While Magnetic Resonance Imaging (MRI) is increasingly used to guide targeted biopsies for prostate cancer diagnosis, its utility remains limited due to high rates of false positives and false negatives as well as low inter-reader agreements. Machine learning methods to detect and localize cancer on prostate MRI can help standardize radiologist interpretations. However, existing machine learning methods vary not only in model architecture, but also in the ground truth labeling strategies used for model training. In this study, we compare different labeling strategies, namely, pathology-confirmed radiologist labels, pathologist labels on whole-mount histopathology images, and lesion-level and pixel-level digital pathologist labels (previously validated deep learning algorithm on histopathology images to predict pixel-level Gleason patterns) on whole-mount histopathology images. We analyse the effects these labels have on the performance of the trained machine learning models. Our experiments show that (1) radiologist labels and models trained with them can miss cancers, or underestimate cancer extent, (2) digital pathologist labels and models trained with them have high concordance with pathologist labels, and (3) models trained with digital pathologist labels achieve the best performance in prostate cancer detection in two different cohorts with different disease distributions, irrespective of the model architecture used. Digital pathologist labels can reduce challenges associated with human annotations, including labor, time, inter- and intra-reader variability, and can help bridge the gap between prostate radiology and pathology by enabling the training of reliable machine learning models to detect and localize prostate cancer on MRI.
Adaptable image quality assessment using meta-reinforcement learning of task amenability
Jul 31, 2021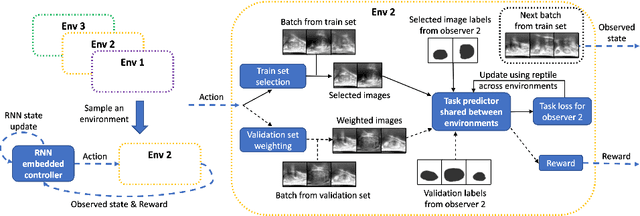
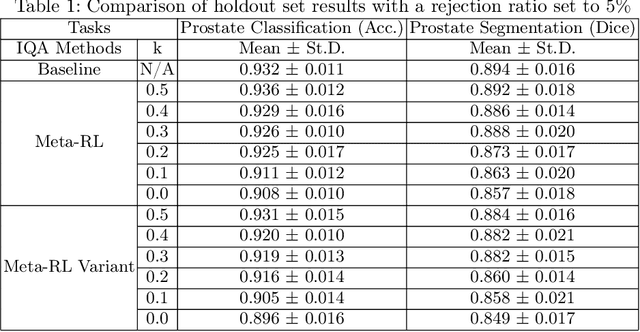
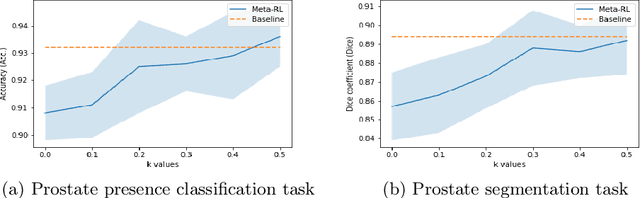
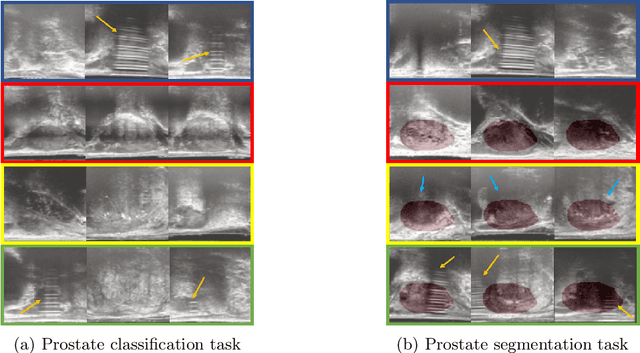
Abstract:The performance of many medical image analysis tasks are strongly associated with image data quality. When developing modern deep learning algorithms, rather than relying on subjective (human-based) image quality assessment (IQA), task amenability potentially provides an objective measure of task-specific image quality. To predict task amenability, an IQA agent is trained using reinforcement learning (RL) with a simultaneously optimised task predictor, such as a classification or segmentation neural network. In this work, we develop transfer learning or adaptation strategies to increase the adaptability of both the IQA agent and the task predictor so that they are less dependent on high-quality, expert-labelled training data. The proposed transfer learning strategy re-formulates the original RL problem for task amenability in a meta-reinforcement learning (meta-RL) framework. The resulting algorithm facilitates efficient adaptation of the agent to different definitions of image quality, each with its own Markov decision process environment including different images, labels and an adaptable task predictor. Our work demonstrates that the IQA agents pre-trained on non-expert task labels can be adapted to predict task amenability as defined by expert task labels, using only a small set of expert labels. Using 6644 clinical ultrasound images from 249 prostate cancer patients, our results for image classification and segmentation tasks show that the proposed IQA method can be adapted using data with as few as respective 19.7% and 29.6% expert-reviewed consensus labels and still achieve comparable IQA and task performance, which would otherwise require a training dataset with 100% expert labels.
Weakly Supervised Registration of Prostate MRI and Histopathology Images
Jun 23, 2021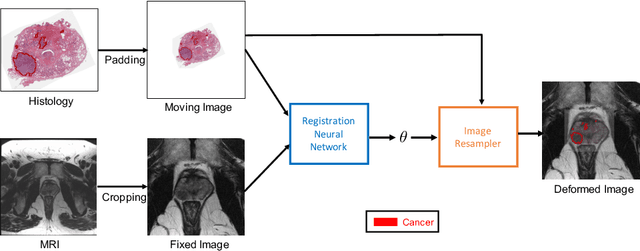
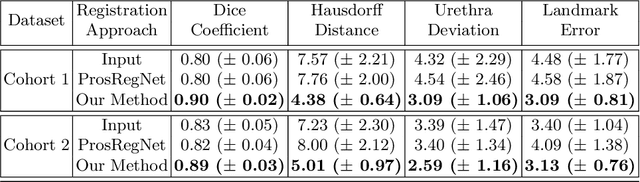
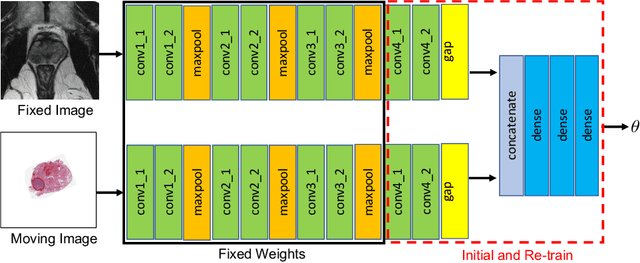
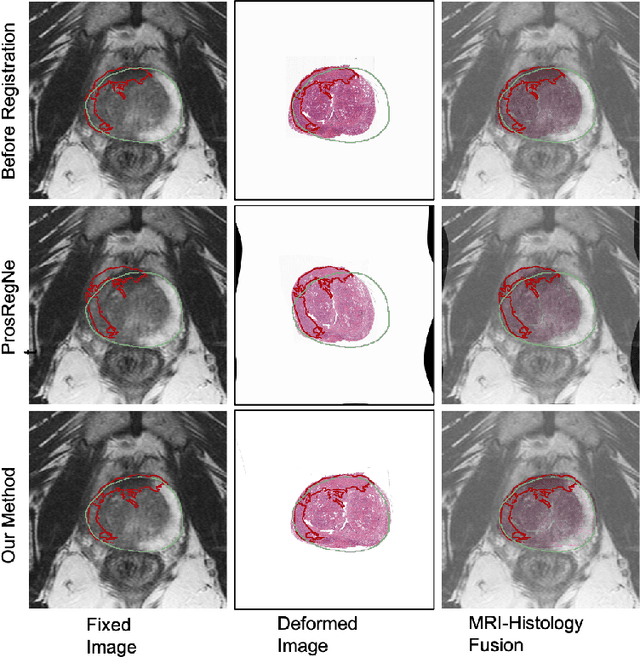
Abstract:The interpretation of prostate MRI suffers from low agreement across radiologists due to the subtle differences between cancer and normal tissue. Image registration addresses this issue by accurately mapping the ground-truth cancer labels from surgical histopathology images onto MRI. Cancer labels achieved by image registration can be used to improve radiologists' interpretation of MRI by training deep learning models for early detection of prostate cancer. A major limitation of current automated registration approaches is that they require manual prostate segmentations, which is a time-consuming task, prone to errors. This paper presents a weakly supervised approach for affine and deformable registration of MRI and histopathology images without requiring prostate segmentations. We used manual prostate segmentations and mono-modal synthetic image pairs to train our registration networks to align prostate boundaries and local prostate features. Although prostate segmentations were used during the training of the network, such segmentations were not needed when registering unseen images at inference time. We trained and validated our registration network with 135 and 10 patients from an internal cohort, respectively. We tested the performance of our method using 16 patients from the internal cohort and 22 patients from an external cohort. The results show that our weakly supervised method has achieved significantly higher registration accuracy than a state-of-the-art method run without prostate segmentations. Our deep learning framework will ease the registration of MRI and histopathology images by obviating the need for prostate segmentations.
Geodesic Density Regression for Correcting 4DCT Pulmonary Respiratory Motion Artifacts
Jun 12, 2021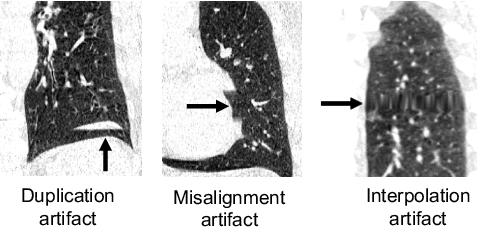
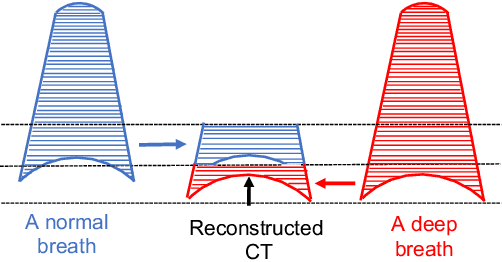
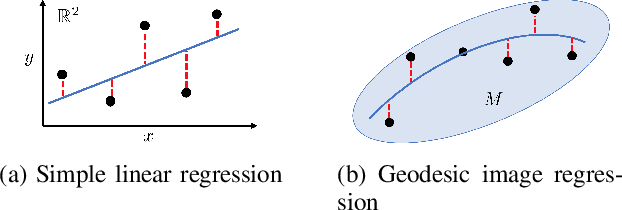
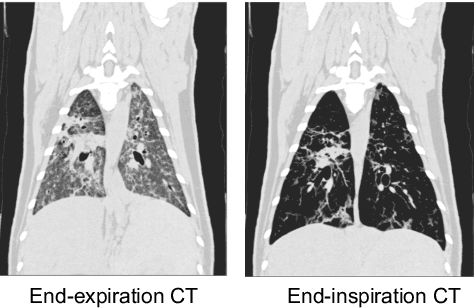
Abstract:Pulmonary respiratory motion artifacts are common in four-dimensional computed tomography (4DCT) of lungs and are caused by missing, duplicated, and misaligned image data. This paper presents a geodesic density regression (GDR) algorithm to correct motion artifacts in 4DCT by correcting artifacts in one breathing phase with artifact-free data from corresponding regions of other breathing phases. The GDR algorithm estimates an artifact-free lung template image and a smooth, dense, 4D (space plus time) vector field that deforms the template image to each breathing phase to produce an artifact-free 4DCT scan. Correspondences are estimated by accounting for the local tissue density change associated with air entering and leaving the lungs, and using binary artifact masks to exclude regions with artifacts from image regression. The artifact-free lung template image is generated by mapping the artifact-free regions of each phase volume to a common reference coordinate system using the estimated correspondences and then averaging. This procedure generates a fixed view of the lung with an improved signal-to-noise ratio. The GDR algorithm was evaluated and compared to a state-of-the-art geodesic intensity regression (GIR) algorithm using simulated CT time-series and 4DCT scans with clinically observed motion artifacts. The simulation shows that the GDR algorithm has achieved significantly more accurate Jacobian images and sharper template images, and is less sensitive to data dropout than the GIR algorithm. We also demonstrate that the GDR algorithm is more effective than the GIR algorithm for removing clinically observed motion artifacts in treatment planning 4DCT scans. Our code is freely available at https://github.com/Wei-Shao-Reg/GDR.
Learning image quality assessment by reinforcing task amenable data selection
Feb 15, 2021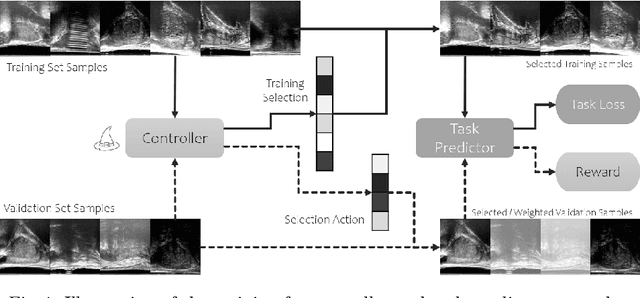
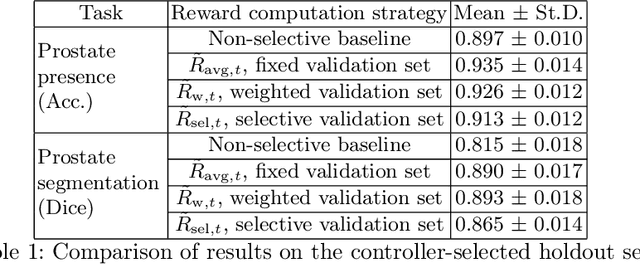
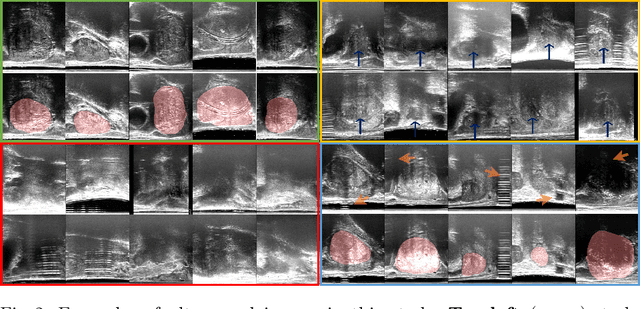
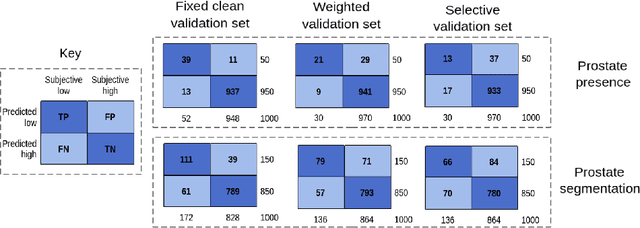
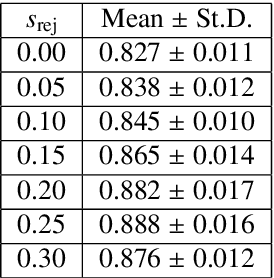
Abstract:In this paper, we consider a type of image quality assessment as a task-specific measurement, which can be used to select images that are more amenable to a given target task, such as image classification or segmentation. We propose to train simultaneously two neural networks for image selection and a target task using reinforcement learning. A controller network learns an image selection policy by maximising an accumulated reward based on the target task performance on the controller-selected validation set, whilst the target task predictor is optimised using the training set. The trained controller is therefore able to reject those images that lead to poor accuracy in the target task. In this work, we show that the controller-predicted image quality can be significantly different from the task-specific image quality labels that are manually defined by humans. Furthermore, we demonstrate that it is possible to learn effective image quality assessment without using a ``clean'' validation set, thereby avoiding the requirement for human labelling of images with respect to their amenability for the task. Using $6712$, labelled and segmented, clinical ultrasound images from $259$ patients, experimental results on holdout data show that the proposed image quality assessment achieved a mean classification accuracy of $0.94\pm0.01$ and a mean segmentation Dice of $0.89\pm0.02$, by discarding $5\%$ and $15\%$ of the acquired images, respectively. The significantly improved performance was observed for both tested tasks, compared with the respective $0.90\pm0.01$ and $0.82\pm0.02$ from networks without considering task amenability. This enables image quality feedback during real-time ultrasound acquisition among many other medical imaging applications.
CorrSigNet: Learning CORRelated Prostate Cancer SIGnatures from Radiology and Pathology Images for Improved Computer Aided Diagnosis
Jul 31, 2020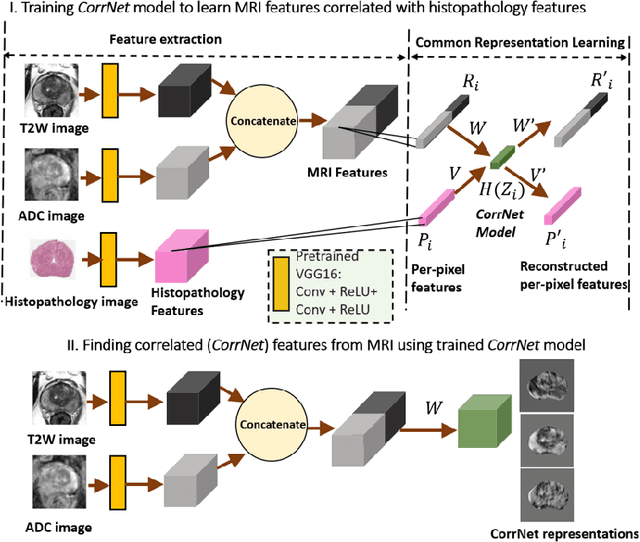
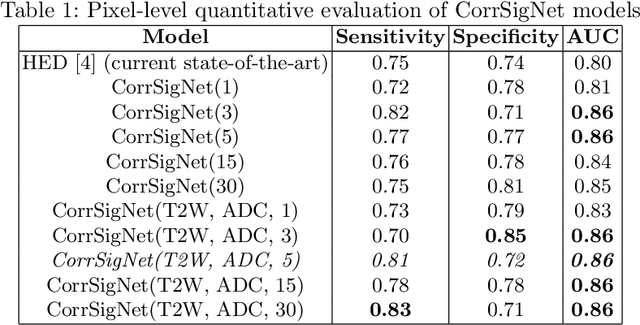
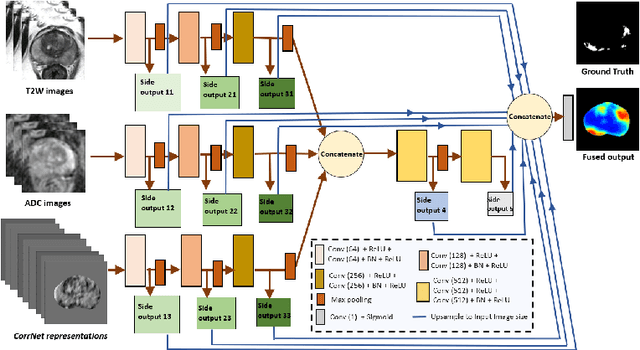
Abstract:Magnetic Resonance Imaging (MRI) is widely used for screening and staging prostate cancer. However, many prostate cancers have subtle features which are not easily identifiable on MRI, resulting in missed diagnoses and alarming variability in radiologist interpretation. Machine learning models have been developed in an effort to improve cancer identification, but current models localize cancer using MRI-derived features, while failing to consider the disease pathology characteristics observed on resected tissue. In this paper, we propose CorrSigNet, an automated two-step model that localizes prostate cancer on MRI by capturing the pathology features of cancer. First, the model learns MRI signatures of cancer that are correlated with corresponding histopathology features using Common Representation Learning. Second, the model uses the learned correlated MRI features to train a Convolutional Neural Network to localize prostate cancer. The histopathology images are used only in the first step to learn the correlated features. Once learned, these correlated features can be extracted from MRI of new patients (without histopathology or surgery) to localize cancer. We trained and validated our framework on a unique dataset of 75 patients with 806 slices who underwent MRI followed by prostatectomy surgery. We tested our method on an independent test set of 20 prostatectomy patients (139 slices, 24 cancerous lesions, 1.12M pixels) and achieved a per-pixel sensitivity of 0.81, specificity of 0.71, AUC of 0.86 and a per-lesion AUC of $0.96 \pm 0.07$, outperforming the current state-of-the-art accuracy in predicting prostate cancer using MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge