Mathias Unberath
Text-Driven Reasoning Video Editing via Reinforcement Learning on Digital Twin Representations
Nov 18, 2025Abstract:Text-driven video editing enables users to modify video content only using text queries. While existing methods can modify video content if explicit descriptions of editing targets with precise spatial locations and temporal boundaries are provided, these requirements become impractical when users attempt to conceptualize edits through implicit queries referencing semantic properties or object relationships. We introduce reasoning video editing, a task where video editing models must interpret implicit queries through multi-hop reasoning to infer editing targets before executing modifications, and a first model attempting to solve this complex task, RIVER (Reasoning-based Implicit Video Editor). RIVER decouples reasoning from generation through digital twin representations of video content that preserve spatial relationships, temporal trajectories, and semantic attributes. A large language model then processes this representation jointly with the implicit query, performing multi-hop reasoning to determine modifications, then outputs structured instructions that guide a diffusion-based editor to execute pixel-level changes. RIVER training uses reinforcement learning with rewards that evaluate reasoning accuracy and generation quality. Finally, we introduce RVEBenchmark, a benchmark of 100 videos with 519 implicit queries spanning three levels and categories of reasoning complexity specifically for reasoning video editing. RIVER demonstrates best performance on the proposed RVEBenchmark and also achieves state-of-the-art performance on two additional video editing benchmarks (VegGIE and FiVE), where it surpasses six baseline methods.
Reasoning Text-to-Video Retrieval via Digital Twin Video Representations and Large Language Models
Nov 15, 2025Abstract:The goal of text-to-video retrieval is to search large databases for relevant videos based on text queries. Existing methods have progressed to handling explicit queries where the visual content of interest is described explicitly; however, they fail with implicit queries where identifying videos relevant to the query requires reasoning. We introduce reasoning text-to-video retrieval, a paradigm that extends traditional retrieval to process implicit queries through reasoning while providing object-level grounding masks that identify which entities satisfy the query conditions. Instead of relying on vision-language models directly, we propose representing video content as digital twins, i.e., structured scene representations that decompose salient objects through specialist vision models. This approach is beneficial because it enables large language models to reason directly over long-horizon video content without visual token compression. Specifically, our two-stage framework first performs compositional alignment between decomposed sub-queries and digital twin representations for candidate identification, then applies large language model-based reasoning with just-in-time refinement that invokes additional specialist models to address information gaps. We construct a benchmark of 447 manually created implicit queries with 135 videos (ReasonT2VBench-135) and another more challenging version of 1000 videos (ReasonT2VBench-1000). Our method achieves 81.2% R@1 on ReasonT2VBench-135, outperforming the strongest baseline by greater than 50 percentage points, and maintains 81.7% R@1 on the extended configuration while establishing state-of-the-art results in three conventional benchmarks (MSR-VTT, MSVD, and VATEX).
Fast Reasoning Segmentation for Images and Videos
Nov 15, 2025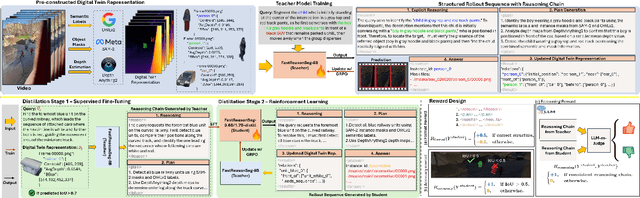
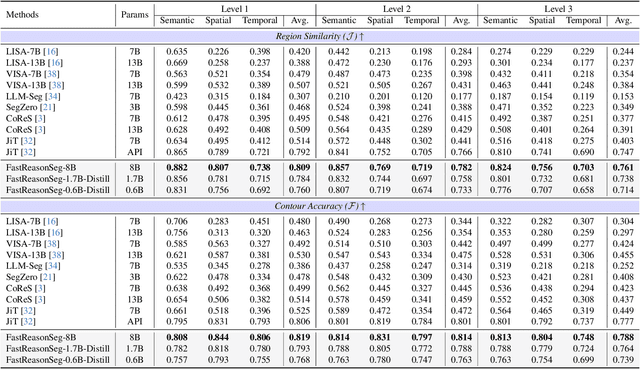
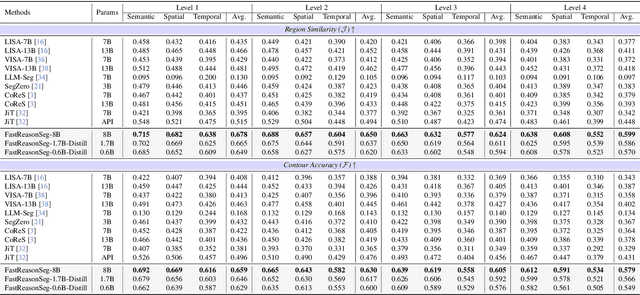

Abstract:Reasoning segmentation enables open-set object segmentation via implicit text queries, therefore serving as a foundation for embodied agents that should operate autonomously in real-world environments. However, existing methods for reasoning segmentation require multimodal large language models with billions of parameters that exceed the computational capabilities of edge devices that typically deploy the embodied AI systems. Distillation offers a pathway to compress these models while preserving their capabilities. Yet, existing distillation approaches fail to transfer the multi-step reasoning capabilities that reasoning segmentation demands, as they focus on matching output predictions and intermediate features rather than preserving reasoning chains. The emerging paradigm of reasoning over digital twin representations presents an opportunity for more effective distillation by re-framing the problem. Consequently, we propose FastReasonSeg, which employs digital twin representations that decouple perception from reasoning to enable more effective distillation. Our distillation scheme first relies on supervised fine-tuning on teacher-generated reasoning chains. Then it is followed by reinforcement fine-tuning with joint rewards evaluating both segmentation accuracy and reasoning quality alignment. Experiments on two video (JiTBench, RVTBench) and two image benchmarks (ReasonSeg, LLM-Seg40K) demonstrate that our FastReasonSeg achieves state-of-the-art reasoning segmentation performance. Moreover, the distilled 0.6B variant outperforms models with 20 times more parameters while achieving 7.79 FPS throughput with only 2.1GB memory consumption. This efficiency enables deployment in resource-constrained environments to enable real-time reasoning segmentation.
Constructing and Interpreting Digital Twin Representations for Visual Reasoning via Reinforcement Learning
Nov 15, 2025Abstract:Visual reasoning may require models to interpret images and videos and respond to implicit text queries across diverse output formats, from pixel-level segmentation masks to natural language descriptions. Existing approaches rely on supervised fine-tuning with task-specific architectures. For example, reasoning segmentation, grounding, summarization, and visual question answering each demand distinct model designs and training, preventing unified solutions and limiting cross-task and cross-modality generalization. Hence, we propose DT-R1, a reinforcement learning framework that trains large language models to construct digital twin representations of complex multi-modal visual inputs and then reason over these high-level representations as a unified approach to visual reasoning. Specifically, we train DT-R1 using GRPO with a novel reward that validates both structural integrity and output accuracy. Evaluations in six visual reasoning benchmarks, covering two modalities and four task types, demonstrate that DT-R1 consistently achieves improvements over state-of-the-art task-specific models. DT-R1 opens a new direction where visual reasoning emerges from reinforcement learning with digital twin representations.
BronchOpt : Vision-Based Pose Optimization with Fine-Tuned Foundation Models for Accurate Bronchoscopy Navigation
Nov 12, 2025



Abstract:Accurate intra-operative localization of the bronchoscope tip relative to patient anatomy remains challenging due to respiratory motion, anatomical variability, and CT-to-body divergence that cause deformation and misalignment between intra-operative views and pre-operative CT. Existing vision-based methods often fail to generalize across domains and patients, leading to residual alignment errors. This work establishes a generalizable foundation for bronchoscopy navigation through a robust vision-based framework and a new synthetic benchmark dataset that enables standardized and reproducible evaluation. We propose a vision-based pose optimization framework for frame-wise 2D-3D registration between intra-operative endoscopic views and pre-operative CT anatomy. A fine-tuned modality- and domain-invariant encoder enables direct similarity computation between real endoscopic RGB frames and CT-rendered depth maps, while a differentiable rendering module iteratively refines camera poses through depth consistency. To enhance reproducibility, we introduce the first public synthetic benchmark dataset for bronchoscopy navigation, addressing the lack of paired CT-endoscopy data. Trained exclusively on synthetic data distinct from the benchmark, our model achieves an average translational error of 2.65 mm and a rotational error of 0.19 rad, demonstrating accurate and stable localization. Qualitative results on real patient data further confirm strong cross-domain generalization, achieving consistent frame-wise 2D-3D alignment without domain-specific adaptation. Overall, the proposed framework achieves robust, domain-invariant localization through iterative vision-based optimization, while the new benchmark provides a foundation for standardized progress in vision-based bronchoscopy navigation.
DualVision ArthroNav: Investigating Opportunities to Enhance Localization and Reconstruction in Image-based Arthroscopy Navigation via External Cameras
Nov 12, 2025


Abstract:Arthroscopic procedures can greatly benefit from navigation systems that enhance spatial awareness, depth perception, and field of view. However, existing optical tracking solutions impose strict workspace constraints and disrupt surgical workflow. Vision-based alternatives, though less invasive, often rely solely on the monocular arthroscope camera, making them prone to drift, scale ambiguity, and sensitivity to rapid motion or occlusion. We propose DualVision ArthroNav, a multi-camera arthroscopy navigation system that integrates an external camera rigidly mounted on the arthroscope. The external camera provides stable visual odometry and absolute localization, while the monocular arthroscope video enables dense scene reconstruction. By combining these complementary views, our system resolves the scale ambiguity and long-term drift inherent in monocular SLAM and ensures robust relocalization. Experiments demonstrate that our system effectively compensates for calibration errors, achieving an average absolute trajectory error of 1.09 mm. The reconstructed scenes reach an average target registration error of 2.16 mm, with high visual fidelity (SSIM = 0.69, PSNR = 22.19). These results indicate that our system provides a practical and cost-efficient solution for arthroscopic navigation, bridging the gap between optical tracking and purely vision-based systems, and paving the way toward clinically deployable, fully vision-based arthroscopic guidance.
TwinOR: Photorealistic Digital Twins of Dynamic Operating Rooms for Embodied AI Research
Nov 10, 2025Abstract:Developing embodied AI for intelligent surgical systems requires safe, controllable environments for continual learning and evaluation. However, safety regulations and operational constraints in operating rooms (ORs) limit embodied agents from freely perceiving and interacting in realistic settings. Digital twins provide high-fidelity, risk-free environments for exploration and training. How we may create photorealistic and dynamic digital representations of ORs that capture relevant spatial, visual, and behavioral complexity remains unclear. We introduce TwinOR, a framework for constructing photorealistic, dynamic digital twins of ORs for embodied AI research. The system reconstructs static geometry from pre-scan videos and continuously models human and equipment motion through multi-view perception of OR activities. The static and dynamic components are fused into an immersive 3D environment that supports controllable simulation and embodied exploration. The proposed framework reconstructs complete OR geometry with centimeter level accuracy while preserving dynamic interaction across surgical workflows, enabling realistic renderings and a virtual playground for embodied AI systems. In our experiments, TwinOR simulates stereo and monocular sensor streams for geometry understanding and visual localization tasks. Models such as FoundationStereo and ORB-SLAM3 on TwinOR-synthesized data achieve performance within their reported accuracy on real indoor datasets, demonstrating that TwinOR provides sensor-level realism sufficient for perception and localization challenges. By establishing a real-to-sim pipeline for constructing dynamic, photorealistic digital twins of OR environments, TwinOR enables the safe, scalable, and data-efficient development and benchmarking of embodied AI, ultimately accelerating the deployment of embodied AI from sim-to-real.
Investigating Robot Control Policy Learning for Autonomous X-ray-guided Spine Procedures
Nov 05, 2025Abstract:Imitation learning-based robot control policies are enjoying renewed interest in video-based robotics. However, it remains unclear whether this approach applies to X-ray-guided procedures, such as spine instrumentation. This is because interpretation of multi-view X-rays is complex. We examine opportunities and challenges for imitation policy learning in bi-plane-guided cannula insertion. We develop an in silico sandbox for scalable, automated simulation of X-ray-guided spine procedures with a high degree of realism. We curate a dataset of correct trajectories and corresponding bi-planar X-ray sequences that emulate the stepwise alignment of providers. We then train imitation learning policies for planning and open-loop control that iteratively align a cannula solely based on visual information. This precisely controlled setup offers insights into limitations and capabilities of this method. Our policy succeeded on the first attempt in 68.5% of cases, maintaining safe intra-pedicular trajectories across diverse vertebral levels. The policy generalized to complex anatomy, including fractures, and remained robust to varied initializations. Rollouts on real bi-planar X-rays further suggest that the model can produce plausible trajectories, despite training exclusively in simulation. While these preliminary results are promising, we also identify limitations, especially in entry point precision. Full closed-look control will require additional considerations around how to provide sufficiently frequent feedback. With more robust priors and domain knowledge, such models may provide a foundation for future efforts toward lightweight and CT-free robotic intra-operative spinal navigation.
Did you just see that? Arbitrary view synthesis for egocentric replay of operating room workflows from ambient sensors
Oct 06, 2025Abstract:Observing surgical practice has historically relied on fixed vantage points or recollections, leaving the egocentric visual perspectives that guide clinical decisions undocumented. Fixed-camera video can capture surgical workflows at the room-scale, but cannot reconstruct what each team member actually saw. Thus, these videos only provide limited insights into how decisions that affect surgical safety, training, and workflow optimization are made. Here we introduce EgoSurg, the first framework to reconstruct the dynamic, egocentric replays for any operating room (OR) staff directly from wall-mounted fixed-camera video, and thus, without intervention to clinical workflow. EgoSurg couples geometry-driven neural rendering with diffusion-based view enhancement, enabling high-visual fidelity synthesis of arbitrary and egocentric viewpoints at any moment. In evaluation across multi-site surgical cases and controlled studies, EgoSurg reconstructs person-specific visual fields and arbitrary viewpoints with high visual quality and fidelity. By transforming existing OR camera infrastructure into a navigable dynamic 3D record, EgoSurg establishes a new foundation for immersive surgical data science, enabling surgical practice to be visualized, experienced, and analyzed from every angle.
Explainable AI for Collaborative Assessment of 2D/3D Registration Quality
Jul 23, 2025Abstract:As surgery embraces digital transformation--integrating sophisticated imaging, advanced algorithms, and robotics to support and automate complex sub-tasks--human judgment of system correctness remains a vital safeguard for patient safety. This shift introduces new "operator-type" roles tasked with verifying complex algorithmic outputs, particularly at critical junctures of the procedure, such as the intermediary check before drilling or implant placement. A prime example is 2D/3D registration, a key enabler of image-based surgical navigation that aligns intraoperative 2D images with preoperative 3D data. Although registration algorithms have advanced significantly, they occasionally yield inaccurate results. Because even small misalignments can lead to revision surgery or irreversible surgical errors, there is a critical need for robust quality assurance. Current visualization-based strategies alone have been found insufficient to enable humans to reliably detect 2D/3D registration misalignments. In response, we propose the first artificial intelligence (AI) framework trained specifically for 2D/3D registration quality verification, augmented by explainability features that clarify the model's decision-making. Our explainable AI (XAI) approach aims to enhance informed decision-making for human operators by providing a second opinion together with a rationale behind it. Through algorithm-centric and human-centered evaluations, we systematically compare four conditions: AI-only, human-only, human-AI, and human-XAI. Our findings reveal that while explainability features modestly improve user trust and willingness to override AI errors, they do not exceed the standalone AI in aggregate performance. Nevertheless, future work extending both the algorithmic design and the human-XAI collaboration elements holds promise for more robust quality assurance of 2D/3D registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge