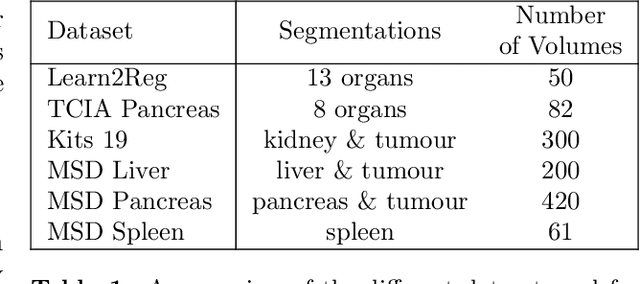
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 08, 2021Alessa Hering, Lasse Hansen, Tony C. W. Mok, Albert C. S. Chung, Hanna Siebert, Stephanie Häger, Annkristin Lange, Sven Kuckertz, Stefan Heldmann, Wei Shao, Sulaiman Vesal, Mirabela Rusu, Geoffrey Sonn, Théo Estienne, Maria Vakalopoulou, Luyi Han, Yunzhi Huang, Mikael Brudfors, Yaël Balbastre, Samuel Joutard, Marc Modat, Gal Lifshitz, Dan Raviv, Jinxin Lv, Qiang Li, Vincent Jaouen, Dimitris Visvikis, Constance Fourcade, Mathieu Rubeaux, Wentao Pan, Zhe Xu, Bailiang Jian, Francesca De Benetti, Marek Wodzinski, Niklas Gunnarsson, Huaqi Qiu, Zeju Li, Christoph Großbröhmer, Andrew Hoopes, Ingerid Reinertsen, Yiming Xiao, Bennett Landman, Yuankai Huo, Keelin Murphy, Bram van Ginneken, Adrian Dalca, Mattias P. Heinrich
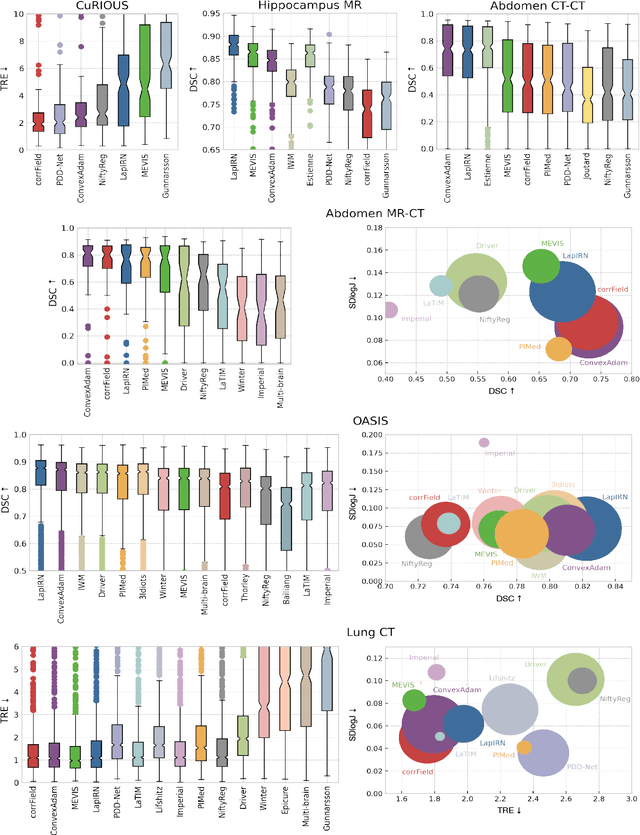
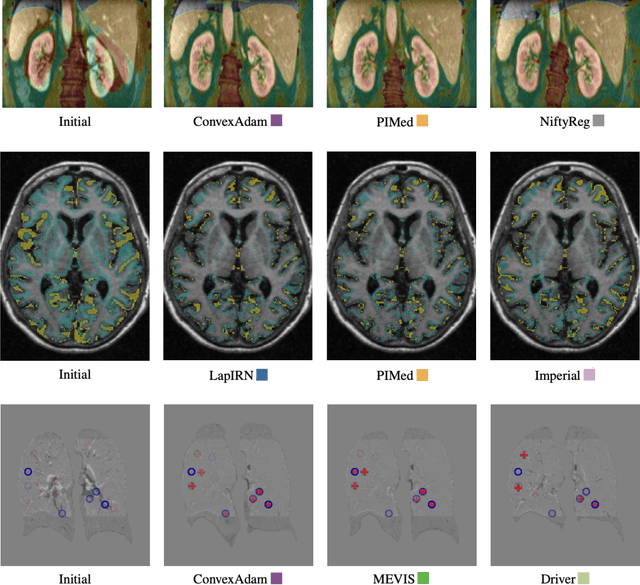
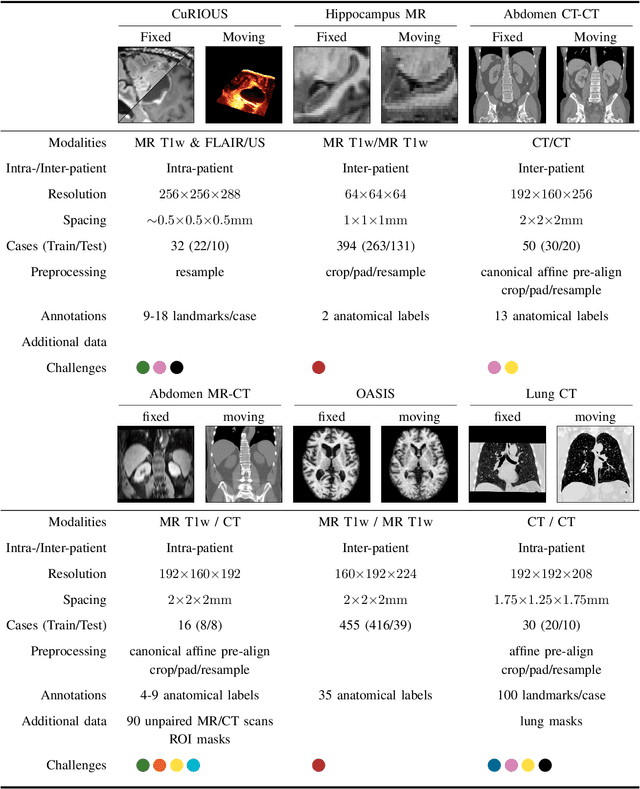
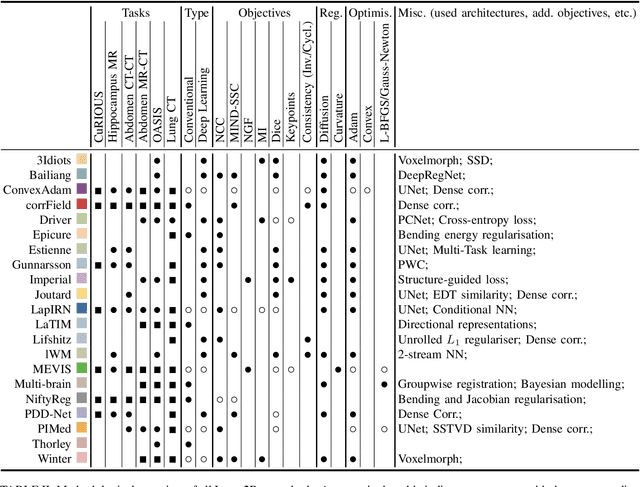
To date few studies have comprehensively compared medical image registration approaches on a wide-range of complementary clinically relevant tasks. This limits the adoption of advances in research into practice and prevents fair benchmarks across competing approaches. Many newer learning-based methods have been explored within the last five years, but the question which optimisation, architectural or metric strategy is ideally suited remains open. Learn2Reg covers a wide range of anatomies: brain, abdomen and thorax, modalities: ultrasound, CT, MRI, populations: intra- and inter-patient and levels of supervision. We established a lower entry barrier for training and validation of 3D registration, which helped us compile results of over 65 individual method submissions from more than 20 unique teams. Our complementary set of metrics, including robustness, accuracy, plausibility and speed enables unique insight into the current-state-of-the-art of medical image registration. Further analyses into transferability, bias and importance of supervision question the superiority of primarily deep learning based approaches and open exiting new research directions into hybrid methods that leverage GPU-accelerated conventional optimisation.
MICS : Multi-steps, Inverse Consistency and Symmetric deep learning registration network
Nov 23, 2021Théo Estienne, Maria Vakalopoulou, Enzo Battistella, Theophraste Henry, Marvin Lerousseau, Amaury Leroy, Nikos Paragios, Eric Deutsch
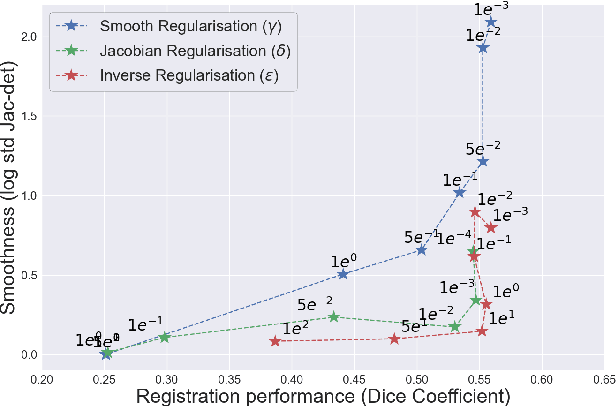

Deformable registration consists of finding the best dense correspondence between two different images. Many algorithms have been published, but the clinical application was made difficult by the high calculation time needed to solve the optimisation problem. Deep learning overtook this limitation by taking advantage of GPU calculation and the learning process. However, many deep learning methods do not take into account desirable properties respected by classical algorithms. In this paper, we present MICS, a novel deep learning algorithm for medical imaging registration. As registration is an ill-posed problem, we focused our algorithm on the respect of different properties: inverse consistency, symmetry and orientation conservation. We also combined our algorithm with a multi-step strategy to refine and improve the deformation grid. While many approaches applied registration to brain MRI, we explored a more challenging body localisation: abdominal CT. Finally, we evaluated our method on a dataset used during the Learn2Reg challenge, allowing a fair comparison with published methods.
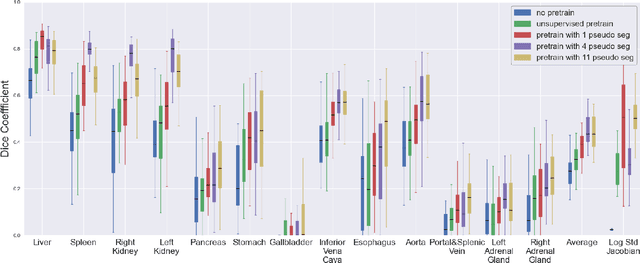
Self-Supervised Representation Learning using Visual Field Expansion on Digital Pathology
Sep 07, 2021Joseph Boyd, Mykola Liashuha, Eric Deutsch, Nikos Paragios, Stergios Christodoulidis, Maria Vakalopoulou
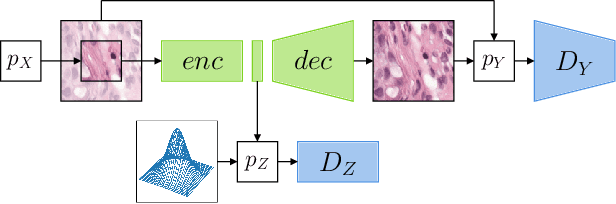
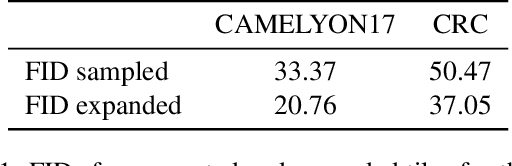
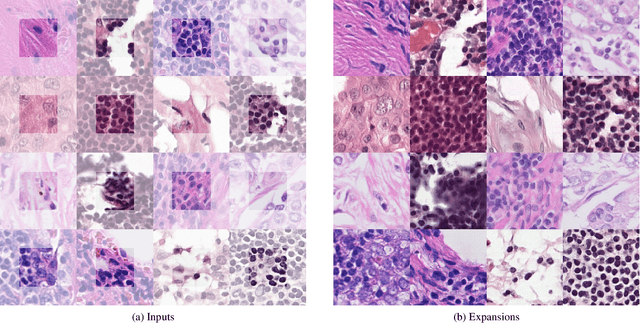
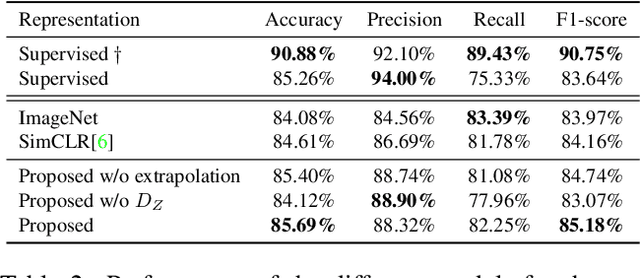
The examination of histopathology images is considered to be the gold standard for the diagnosis and stratification of cancer patients. A key challenge in the analysis of such images is their size, which can run into the gigapixels and can require tedious screening by clinicians. With the recent advances in computational medicine, automatic tools have been proposed to assist clinicians in their everyday practice. Such tools typically process these large images by slicing them into tiles that can then be encoded and utilized for different clinical models. In this study, we propose a novel generative framework that can learn powerful representations for such tiles by learning to plausibly expand their visual field. In particular, we developed a progressively grown generative model with the objective of visual field expansion. Thus trained, our model learns to generate different tissue types with fine details, while simultaneously learning powerful representations that can be used for different clinical endpoints, all in a self-supervised way. To evaluate the performance of our model, we conducted classification experiments on CAMELYON17 and CRC benchmark datasets, comparing favorably to other self-supervised and pre-trained strategies that are commonly used in digital pathology. Our code is available at https://github.com/jcboyd/cdpath21-gan.
Deep Reinforcement Learning for L3 Slice Localization in Sarcopenia Assessment
Aug 13, 2021Othmane Laousy, Guillaume Chassagnon, Edouard Oyallon, Nikos Paragios, Marie-Pierre Revel, Maria Vakalopoulou
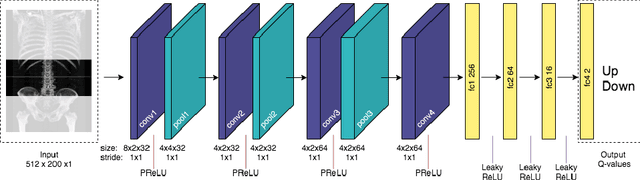
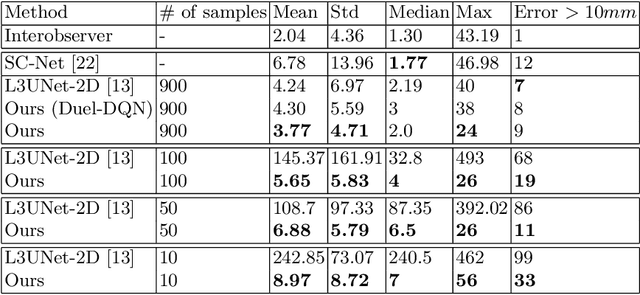
Sarcopenia is a medical condition characterized by a reduction in muscle mass and function. A quantitative diagnosis technique consists of localizing the CT slice passing through the middle of the third lumbar area (L3) and segmenting muscles at this level. In this paper, we propose a deep reinforcement learning method for accurate localization of the L3 CT slice. Our method trains a reinforcement learning agent by incentivizing it to discover the right position. Specifically, a Deep Q-Network is trained to find the best policy to follow for this problem. Visualizing the training process shows that the agent mimics the scrolling of an experienced radiologist. Extensive experiments against other state-of-the-art deep learning based methods for L3 localization prove the superiority of our technique which performs well even with a limited amount of data and annotations.
Exploring Deep Registration Latent Spaces
Jul 23, 2021Théo Estienne, Maria Vakalopoulou, Stergios Christodoulidis, Enzo Battistella, Théophraste Henry, Marvin Lerousseau, Amaury Leroy, Guillaume Chassagnon, Marie-Pierre Revel, Nikos Paragios, Eric Deutsch
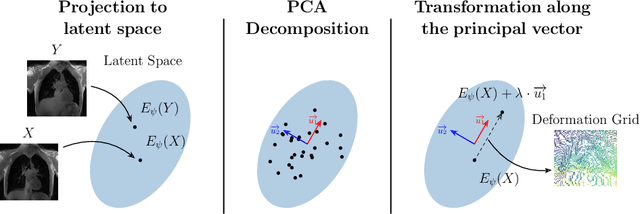
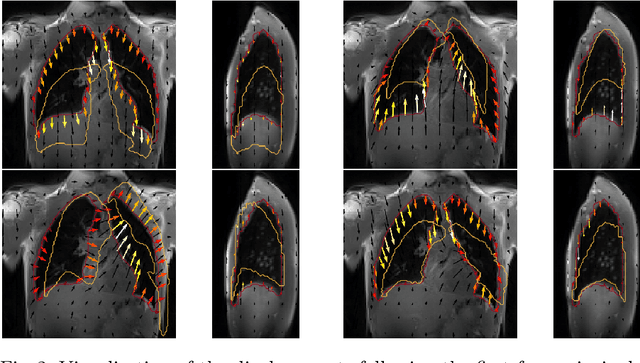
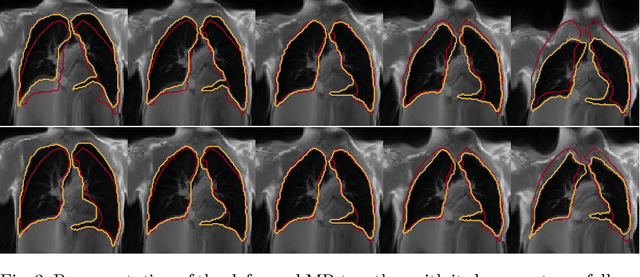
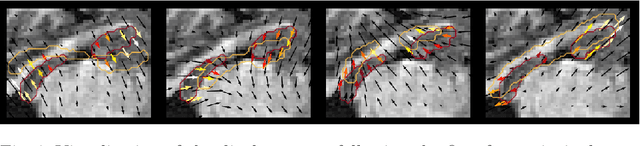
Explainability of deep neural networks is one of the most challenging and interesting problems in the field. In this study, we investigate the topic focusing on the interpretability of deep learning-based registration methods. In particular, with the appropriate model architecture and using a simple linear projection, we decompose the encoding space, generating a new basis, and we empirically show that this basis captures various decomposed anatomically aware geometrical transformations. We perform experiments using two different datasets focusing on lungs and hippocampus MRI. We show that such an approach can decompose the highly convoluted latent spaces of registration pipelines in an orthogonal space with several interesting properties. We hope that this work could shed some light on a better understanding of deep learning-based registration methods.
Multi-Source domain adaptation via supervised contrastive learning and confident consistency regularization
Jul 01, 2021Marin Scalbert, Maria Vakalopoulou, Florent Couzinié-Devy
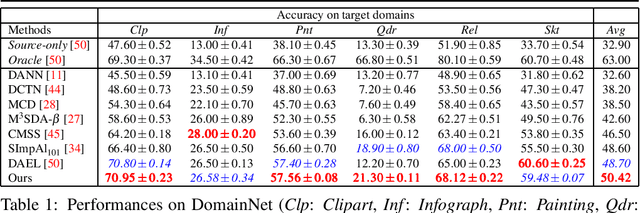
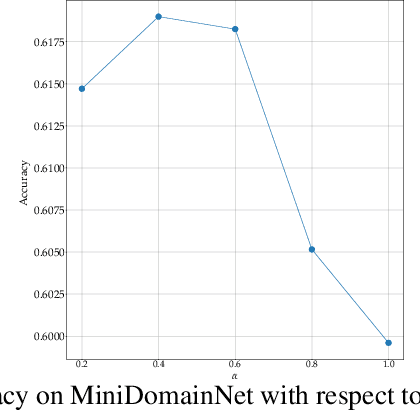
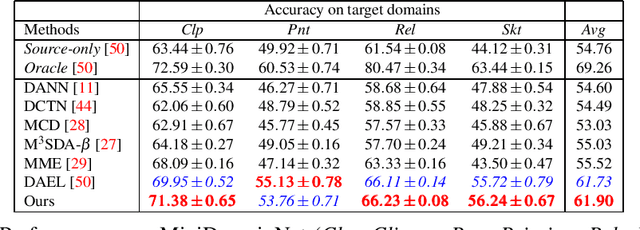
Multi-Source Unsupervised Domain Adaptation (multi-source UDA) aims to learn a model from several labeled source domains while performing well on a different target domain where only unlabeled data are available at training time. To align source and target features distributions, several recent works use source and target explicit statistics matching such as features moments or class centroids. Yet, these approaches do not guarantee class conditional distributions alignment across domains. In this work, we propose a new framework called Contrastive Multi-Source Domain Adaptation (CMSDA) for multi-source UDA that addresses this limitation. Discriminative features are learned from interpolated source examples via cross entropy minimization and from target examples via consistency regularization and hard pseudo-labeling. Simultaneously, interpolated source examples are leveraged to align source class conditional distributions through an interpolated version of the supervised contrastive loss. This alignment leads to more general and transferable features which further improve the generalization on the target domain. Extensive experiments have been carried out on three standard multi-source UDA datasets where our method reports state-of-the-art results.
Weakly supervised pan-cancer segmentation tool
May 10, 2021Marvin Lerousseau, Marion Classe, Enzo Battistella, Théo Estienne, Théophraste Henry, Amaury Leroy, Roger Sun, Maria Vakalopoulou, Jean-Yves Scoazec, Eric Deutsch, Nikos Paragios
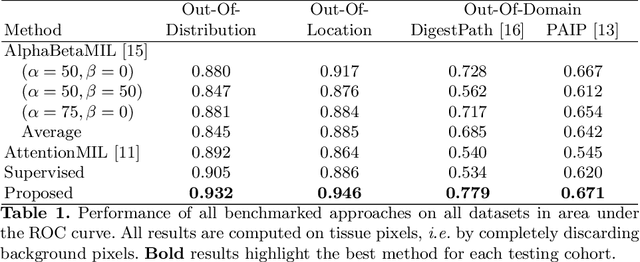

The vast majority of semantic segmentation approaches rely on pixel-level annotations that are tedious and time consuming to obtain and suffer from significant inter and intra-expert variability. To address these issues, recent approaches have leveraged categorical annotations at the slide-level, that in general suffer from robustness and generalization. In this paper, we propose a novel weakly supervised multi-instance learning approach that deciphers quantitative slide-level annotations which are fast to obtain and regularly present in clinical routine. The extreme potentials of the proposed approach are demonstrated for tumor segmentation of solid cancer subtypes. The proposed approach achieves superior performance in out-of-distribution, out-of-location, and out-of-domain testing sets.
Sparse convolutional context-aware multiple instance learning for whole slide image classification
May 06, 2021Marvin Lerousseau, Maria Vakalopoulou, Nikos Paragios, Eric Deutsch
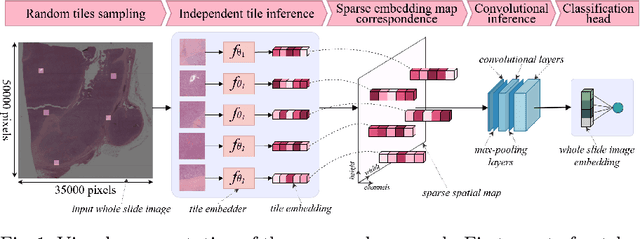
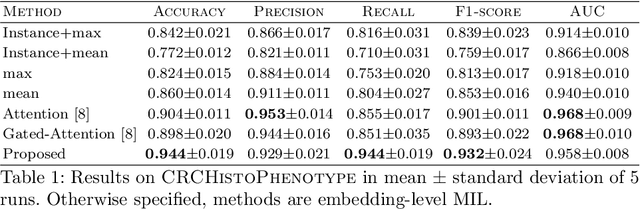
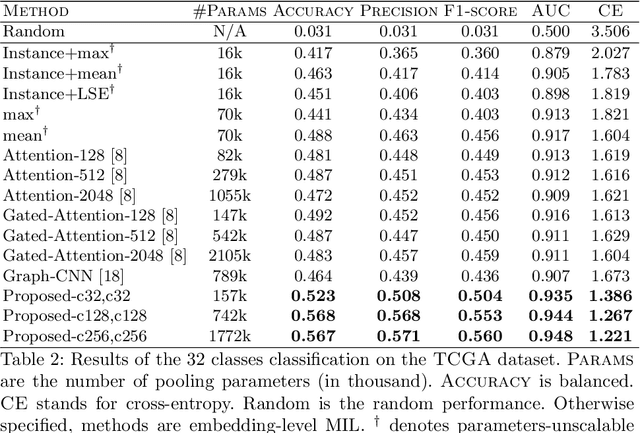
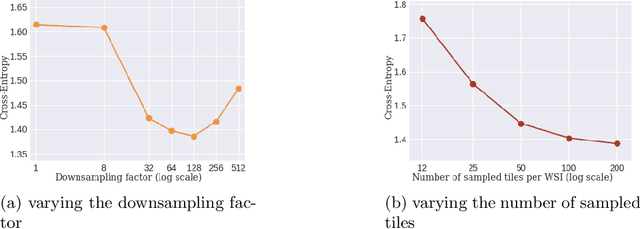
Whole slide microscopic slides display many cues about the underlying tissue guiding diagnostic and the choice of therapy for many diseases. However, their enormous size often in gigapixels hampers the use of traditional neural network architectures. To tackle this issue, multiple instance learning (MIL) classifies bags of patches instead of whole slide images. Most MIL strategies consider that patches are independent and identically distributed. Our approach presents a paradigm shift through the integration of spatial information of patches with a sparse-input convolutional-based MIL strategy. The formulated framework is generic, flexible, scalable and is the first to introduce contextual dependencies between decisions taken at the patch level. It achieved state-of-the-art performance in pan-cancer subtype classification. The code of this work will be made available.
Cancer Gene Profiling through Unsupervised Discovery
Feb 11, 2021Enzo Battistella, Maria Vakalopoulou, Roger Sun, Théo Estienne, Marvin Lerousseau, Sergey Nikolaev, Emilie Alvarez Andres, Alexandre Carré, Stéphane Niyoteka, Charlotte Robert, Nikos Paragios, Eric Deutsch
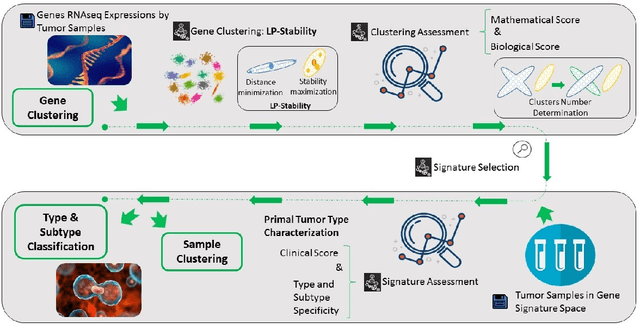
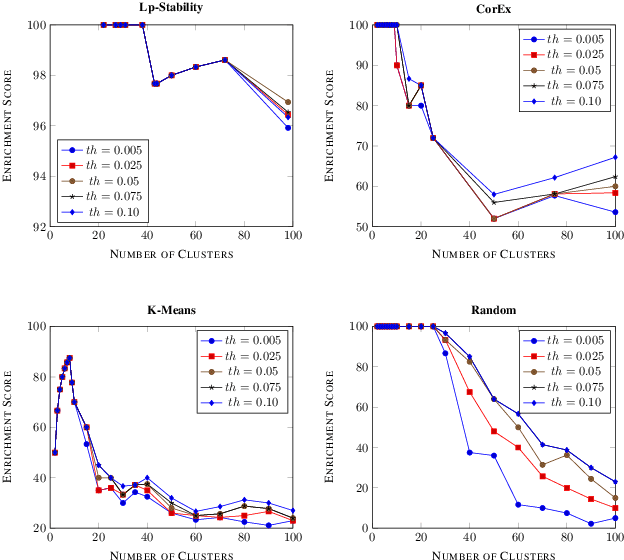
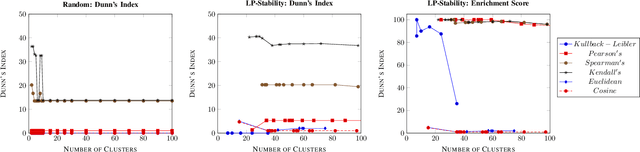
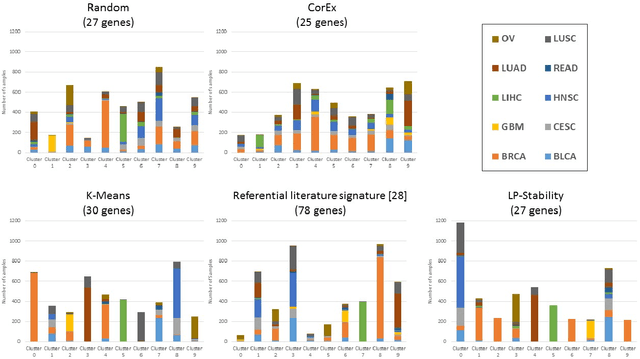
Precision medicine is a paradigm shift in healthcare relying heavily on genomics data. However, the complexity of biological interactions, the large number of genes as well as the lack of comparisons on the analysis of data, remain a tremendous bottleneck regarding clinical adoption. In this paper, we introduce a novel, automatic and unsupervised framework to discover low-dimensional gene biomarkers. Our method is based on the LP-Stability algorithm, a high dimensional center-based unsupervised clustering algorithm, that offers modularity as concerns metric functions and scalability, while being able to automatically determine the best number of clusters. Our evaluation includes both mathematical and biological criteria. The recovered signature is applied to a variety of biological tasks, including screening of biological pathways and functions, and characterization relevance on tumor types and subtypes. Quantitative comparisons among different distance metrics, commonly used clustering methods and a referential gene signature used in the literature, confirm state of the art performance of our approach. In particular, our signature, that is based on 27 genes, reports at least $30$ times better mathematical significance (average Dunn's Index) and 25% better biological significance (average Enrichment in Protein-Protein Interaction) than those produced by other referential clustering methods. Finally, our signature reports promising results on distinguishing immune inflammatory and immune desert tumors, while reporting a high balanced accuracy of 92% on tumor types classification and averaged balanced accuracy of 68% on tumor subtypes classification, which represents, respectively 7% and 9% higher performance compared to the referential signature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge