Matthew F. Glasser
From Low Field to High Value: Robust Cortical Mapping from Low-Field MRI
May 18, 2025Abstract:Three-dimensional reconstruction of cortical surfaces from MRI for morphometric analysis is fundamental for understanding brain structure. While high-field MRI (HF-MRI) is standard in research and clinical settings, its limited availability hinders widespread use. Low-field MRI (LF-MRI), particularly portable systems, offers a cost-effective and accessible alternative. However, existing cortical surface analysis tools are optimized for high-resolution HF-MRI and struggle with the lower signal-to-noise ratio and resolution of LF-MRI. In this work, we present a machine learning method for 3D reconstruction and analysis of portable LF-MRI across a range of contrasts and resolutions. Our method works "out of the box" without retraining. It uses a 3D U-Net trained on synthetic LF-MRI to predict signed distance functions of cortical surfaces, followed by geometric processing to ensure topological accuracy. We evaluate our method using paired HF/LF-MRI scans of the same subjects, showing that LF-MRI surface reconstruction accuracy depends on acquisition parameters, including contrast type (T1 vs T2), orientation (axial vs isotropic), and resolution. A 3mm isotropic T2-weighted scan acquired in under 4 minutes, yields strong agreement with HF-derived surfaces: surface area correlates at r=0.96, cortical parcellations reach Dice=0.98, and gray matter volume achieves r=0.93. Cortical thickness remains more challenging with correlations up to r=0.70, reflecting the difficulty of sub-mm precision with 3mm voxels. We further validate our method on challenging postmortem LF-MRI, demonstrating its robustness. Our method represents a step toward enabling cortical surface analysis on portable LF-MRI. Code is available at https://surfer.nmr.mgh.harvard.edu/fswiki/ReconAny
Surface Vision Transformers: Flexible Attention-Based Modelling of Biomedical Surfaces
Apr 07, 2022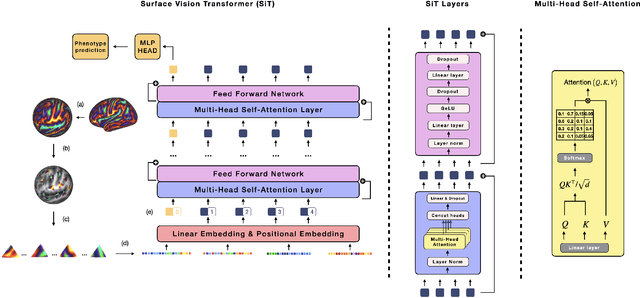
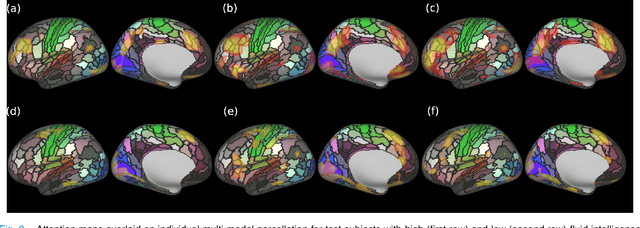
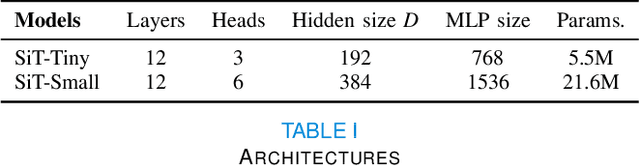
Abstract:Recent state-of-the-art performances of Vision Transformers (ViT) in computer vision tasks demonstrate that a general-purpose architecture, which implements long-range self-attention, could replace the local feature learning operations of convolutional neural networks. In this paper, we extend ViTs to surfaces by reformulating the task of surface learning as a sequence-to-sequence learning problem, by proposing patching mechanisms for general surface meshes. Sequences of patches are then processed by a transformer encoder and used for classification or regression. We validate our method on a range of different biomedical surface domains and tasks: brain age prediction in the developing Human Connectome Project (dHCP), fluid intelligence prediction in the Human Connectome Project (HCP), and coronary artery calcium score classification using surfaces from the Scottish Computed Tomography of the Heart (SCOT-HEART) dataset, and investigate the impact of pretraining and data augmentation on model performance. Results suggest that Surface Vision Transformers (SiT) demonstrate consistent improvement over geometric deep learning methods for brain age and fluid intelligence prediction and achieve comparable performance on calcium score classification to standard metrics used in clinical practice. Furthermore, analysis of transformer attention maps offers clear and individualised predictions of the features driving each task. Code is available on Github: https://github.com/metrics-lab/surface-vision-transformers
Surface Vision Transformers: Attention-Based Modelling applied to Cortical Analysis
Mar 30, 2022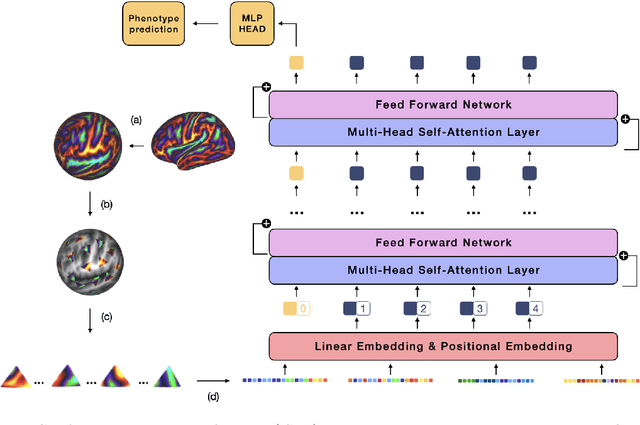

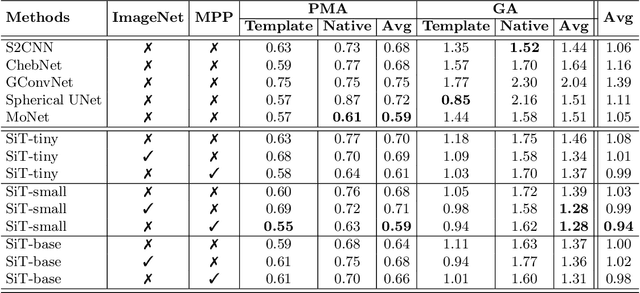
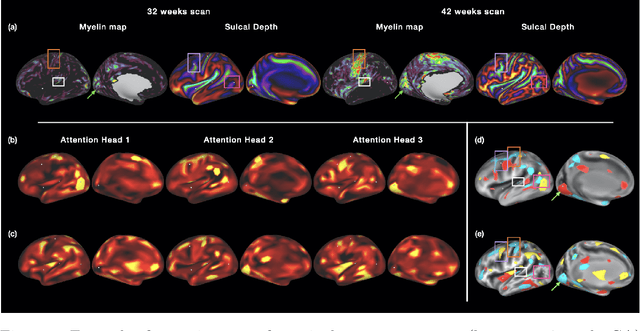
Abstract:The extension of convolutional neural networks (CNNs) to non-Euclidean geometries has led to multiple frameworks for studying manifolds. Many of those methods have shown design limitations resulting in poor modelling of long-range associations, as the generalisation of convolutions to irregular surfaces is non-trivial. Motivated by the success of attention-modelling in computer vision, we translate convolution-free vision transformer approaches to surface data, to introduce a domain-agnostic architecture to study any surface data projected onto a spherical manifold. Here, surface patching is achieved by representing spherical data as a sequence of triangular patches, extracted from a subdivided icosphere. A transformer model encodes the sequence of patches via successive multi-head self-attention layers while preserving the sequence resolution. We validate the performance of the proposed Surface Vision Transformer (SiT) on the task of phenotype regression from cortical surface metrics derived from the Developing Human Connectome Project (dHCP). Experiments show that the SiT generally outperforms surface CNNs, while performing comparably on registered and unregistered data. Analysis of transformer attention maps offers strong potential to characterise subtle cognitive developmental patterns.
ICAM-reg: Interpretable Classification and Regression with Feature Attribution for Mapping Neurological Phenotypes in Individual Scans
Mar 03, 2021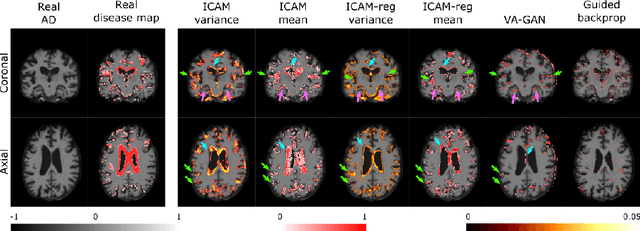
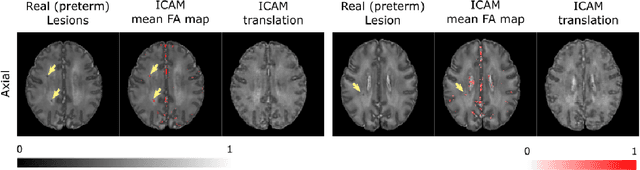
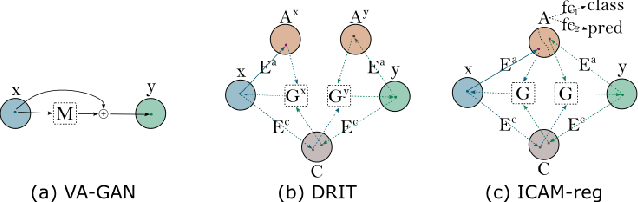
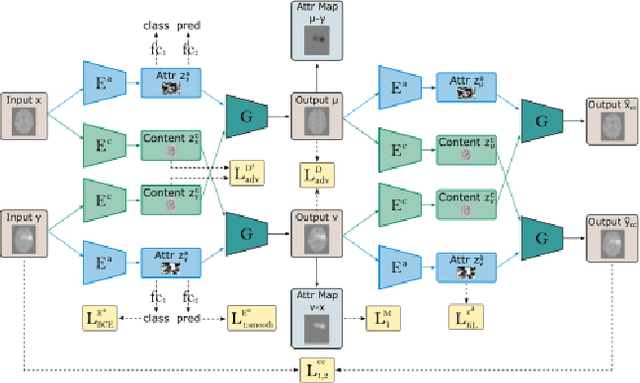
Abstract:An important goal of medical imaging is to be able to precisely detect patterns of disease specific to individual scans; however, this is challenged in brain imaging by the degree of heterogeneity of shape and appearance. Traditional methods, based on image registration to a global template, historically fail to detect variable features of disease, as they utilise population-based analyses, suited primarily to studying group-average effects. In this paper we therefore take advantage of recent developments in generative deep learning to develop a method for simultaneous classification, or regression, and feature attribution (FA). Specifically, we explore the use of a VAE-GAN translation network called ICAM, to explicitly disentangle class relevant features from background confounds for improved interpretability and regression of neurological phenotypes. We validate our method on the tasks of Mini-Mental State Examination (MMSE) cognitive test score prediction for the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort, as well as brain age prediction, for both neurodevelopment and neurodegeneration, using the developing Human Connectome Project (dHCP) and UK Biobank datasets. We show that the generated FA maps can be used to explain outlier predictions and demonstrate that the inclusion of a regression module improves the disentanglement of the latent space. Our code is freely available on Github https://github.com/CherBass/ICAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge