Brian L. Edlow
JParc: Joint cortical surface parcellation with registration
Dec 27, 2025Abstract:Cortical surface parcellation is a fundamental task in both basic neuroscience research and clinical applications, enabling more accurate mapping of brain regions. Model-based and learning-based approaches for automated parcellation alleviate the need for manual labeling. Despite the advancement in parcellation performance, learning-based methods shift away from registration and atlas propagation without exploring the reason for the improvement compared to traditional methods. In this study, we present JParc, a joint cortical registration and parcellation framework, that outperforms existing state-of-the-art parcellation methods. In rigorous experiments, we demonstrate that the enhanced performance of JParc is primarily attributable to accurate cortical registration and a learned parcellation atlas. By leveraging a shallow subnetwork to fine-tune the propagated atlas labels, JParc achieves a Dice score greater than 90% on the Mindboggle dataset, using only basic geometric features (sulcal depth, curvature) that describe cortical folding patterns. The superior accuracy of JParc can significantly increase the statistical power in brain mapping studies as well as support applications in surgical planning and many other downstream neuroscientific and clinical tasks.
Joint cortical registration of geometry and function using semi-supervised learning
Mar 06, 2023Abstract:Brain surface-based image registration, an important component of brain image analysis, establishes spatial correspondence between cortical surfaces. Existing iterative and learning-based approaches focus on accurate registration of folding patterns of the cerebral cortex, and assume that geometry predicts function and thus functional areas will also be well aligned. However, structure/functional variability of anatomically corresponding areas across subjects has been widely reported. In this work, we introduce a learning-based cortical registration framework, JOSA, which jointly aligns folding patterns and functional maps while simultaneously learning an optimal atlas. We demonstrate that JOSA can substantially improve registration performance in both anatomical and functional domains over existing methods. By employing a semi-supervised training strategy, the proposed framework obviates the need for functional data during inference, enabling its use in broad neuroscientific domains where functional data may not be observed.
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast
Dec 24, 2020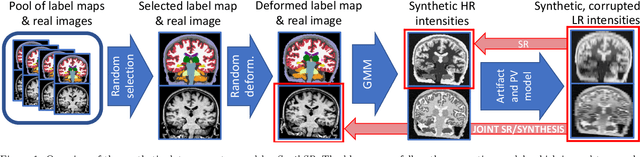

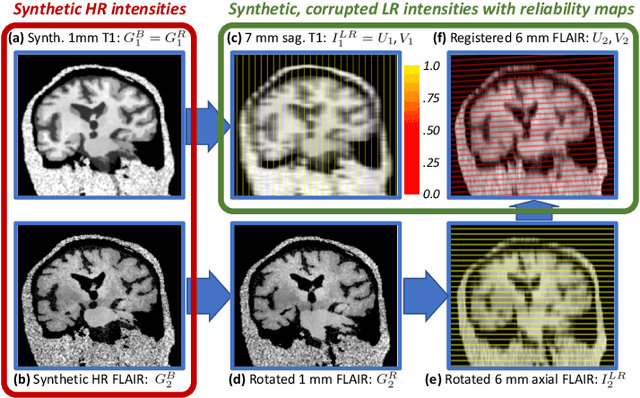
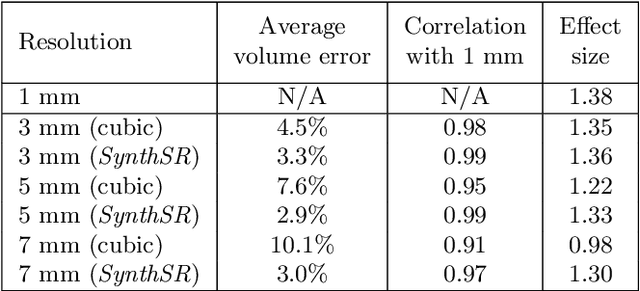
Abstract:Most existing algorithms for automatic 3D morphometry of human brain MRI scans are designed for data with near-isotropic voxels at approximately 1 mm resolution, and frequently have contrast constraints as well - typically requiring T1 scans (e.g., MP-RAGE). This limitation prevents the analysis of millions of MRI scans acquired with large inter-slice spacing ("thick slice") in clinical settings every year. The inability to quantitatively analyze these scans hinders the adoption of quantitative neuroimaging in healthcare, and precludes research studies that could attain huge sample sizes and hence greatly improve our understanding of the human brain. Recent advances in CNNs are producing outstanding results in super-resolution and contrast synthesis of MRI. However, these approaches are very sensitive to the contrast, resolution and orientation of the input images, and thus do not generalize to diverse clinical acquisition protocols - even within sites. Here we present SynthSR, a method to train a CNN that receives one or more thick-slice scans with different contrast, resolution and orientation, and produces an isotropic scan of canonical contrast (typically a 1 mm MP-RAGE). The presented method does not require any preprocessing, e.g., skull stripping or bias field correction. Crucially, SynthSR trains on synthetic input images generated from 3D segmentations, and can thus be used to train CNNs for any combination of contrasts, resolutions and orientations without high-resolution training data. We test the images generated with SynthSR in an array of common downstream analyses, and show that they can be reliably used for subcortical segmentation and volumetry, image registration (e.g., for tensor-based morphometry), and, if some image quality requirements are met, even cortical thickness morphometry. The source code is publicly available at github.com/BBillot/SynthSR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge