Irina Voiculescu
Negational Symmetry of Quantum Neural Networks for Binary Pattern Classification
May 20, 2021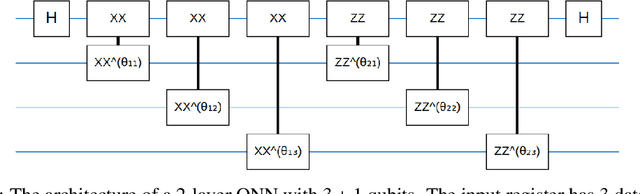
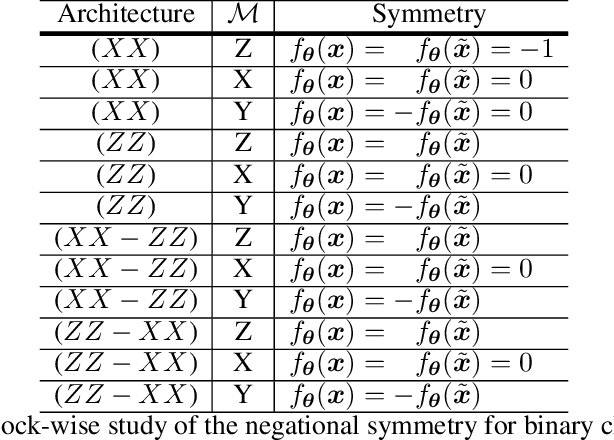
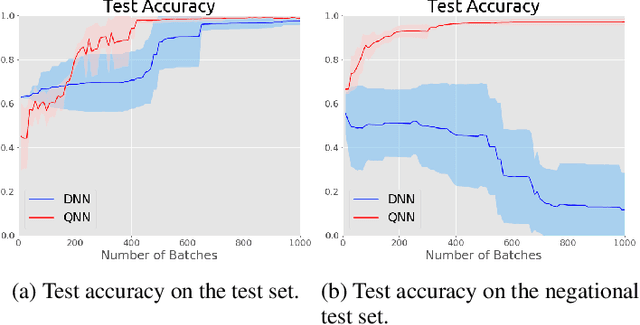
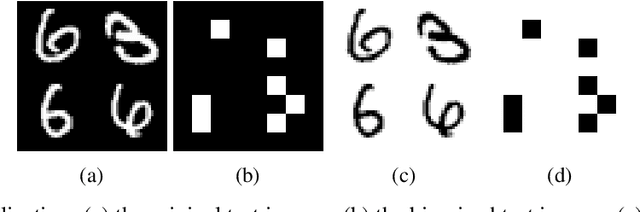
Abstract:Entanglement is a physical phenomenon, which has fueled recent successes of quantum algorithms. Although quantum neural networks (QNNs) have shown promising results in solving simple machine learning tasks recently, for the time being, the effect of entanglement in QNNs and the behavior of QNNs in binary pattern classification are still underexplored. In this work, we provide some theoretical insight into the properties of QNNs by presenting and analyzing a new form of invariance embedded in QNNs for both quantum binary classification and quantum representation learning, which we term negational symmetry. Given a quantum binary signal and its negational counterpart where a bitwise NOT operation is applied to each quantum bit of the binary signal, a QNN outputs the same logits. That is to say, QNNs cannot differentiate a quantum binary signal and its negational counterpart in a binary classification task. We further empirically evaluate the negational symmetry of QNNs in binary pattern classification tasks using Google's quantum computing framework. The theoretical and experimental results suggest that negational symmetry is a fundamental property of QNNs, which is not shared by classical models. Our findings also imply that negational symmetry is a double-edged sword in practical quantum applications.
Towards Robust Medical Image Segmentation on Small-Scale Data with Incomplete Labels
Nov 28, 2020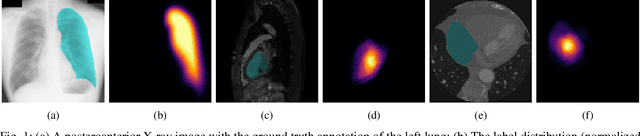
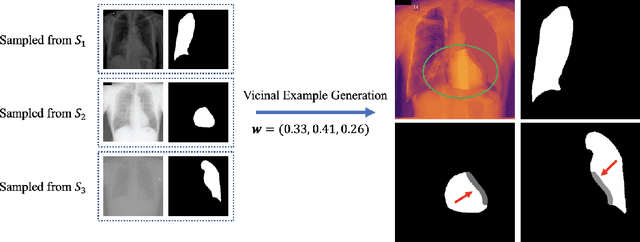
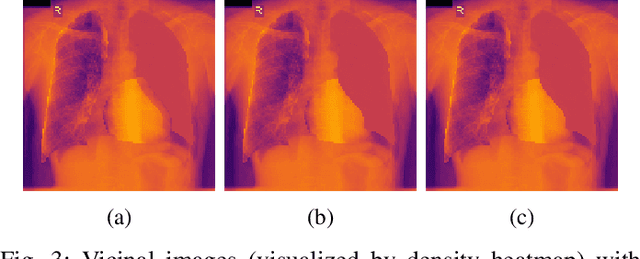
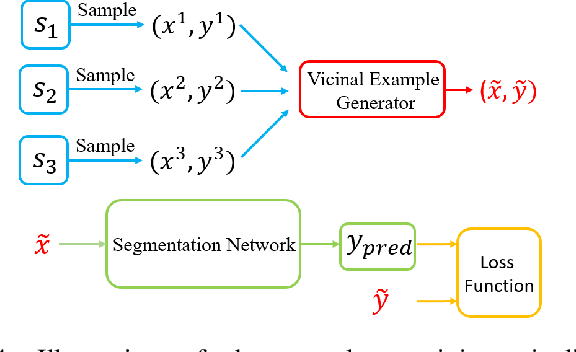
Abstract:The data-driven nature of deep learning models for semantic segmentation requires a large number of pixel-level annotations. However, large-scale and fully labeled medical datasets are often unavailable for practical tasks. Recently, partially supervised methods have been proposed to utilize images with incomplete labels to mitigate the data scarcity problem in the medical domain. As an emerging research area, the breakthroughs made by existing methods rely on either large-scale data or complex model design, which makes them 1) less practical for certain real-life tasks and 2) less robust for small-scale data. It is time to step back and think about the robustness of partially supervised methods and how to maximally utilize small-scale and partially labeled data for medical image segmentation tasks. To bridge the methodological gaps in label-efficient deep learning with partial supervision, we propose RAMP, a simple yet efficient data augmentation framework for partially supervised medical image segmentation by exploiting the assumption that patients share anatomical similarities. We systematically evaluate RAMP and the previous methods in various controlled multi-structure segmentation tasks. Compared to the mainstream approaches, RAMP consistently improves the performance of traditional segmentation networks on small-scale partially labeled data and utilize additional image-wise weak annotations.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020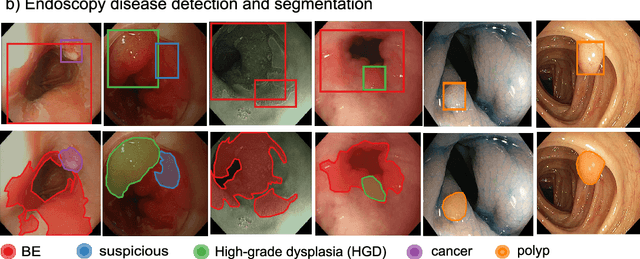
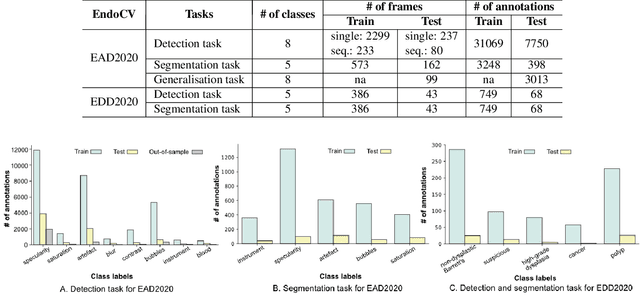
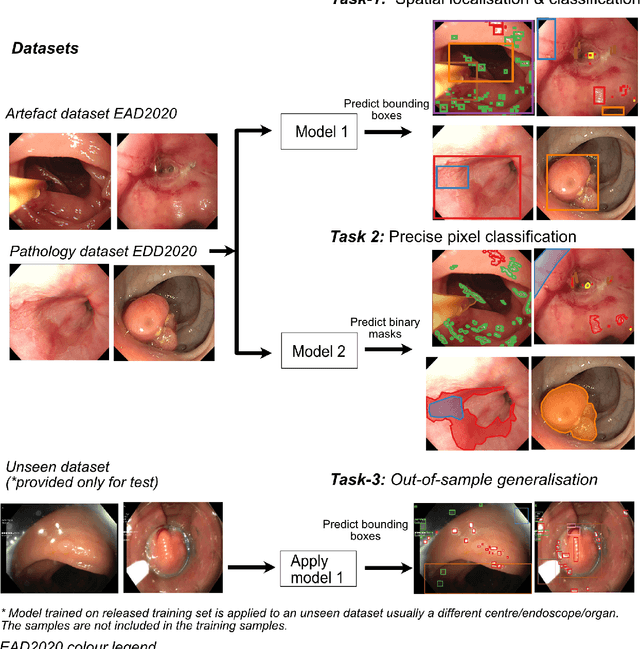
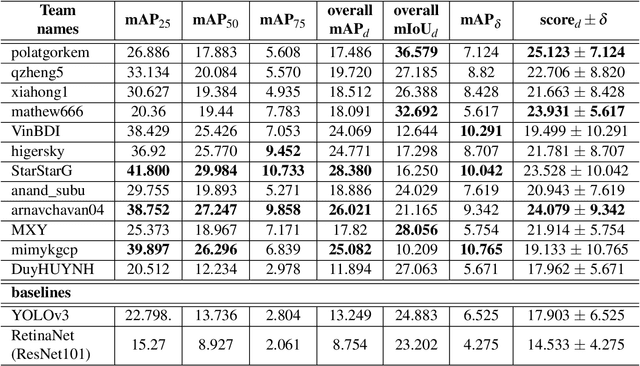
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
RRA-U-Net: a Residual Encoder to Attention Decoder by Residual Connections Framework for Spine Segmentation under Noisy Labels
Sep 27, 2020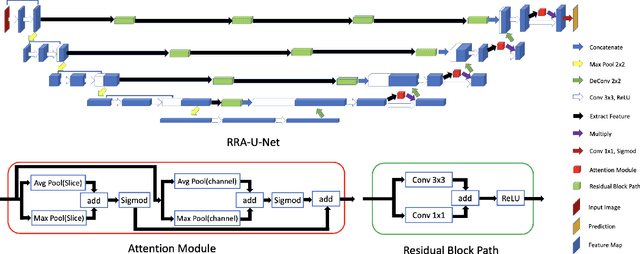
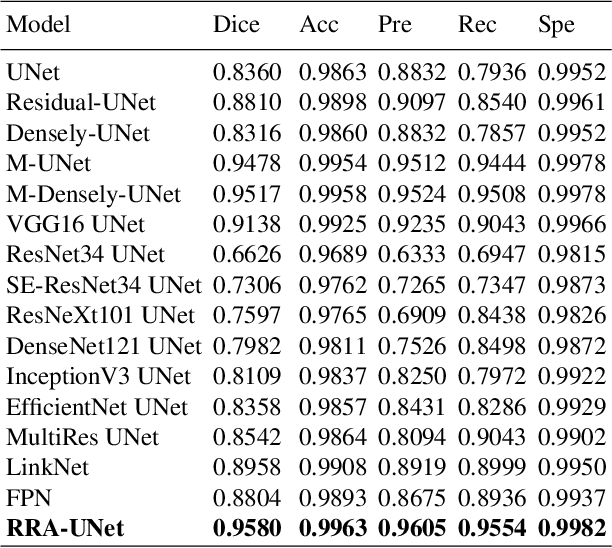
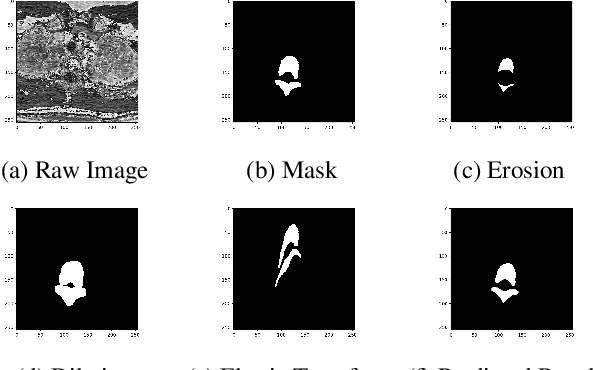
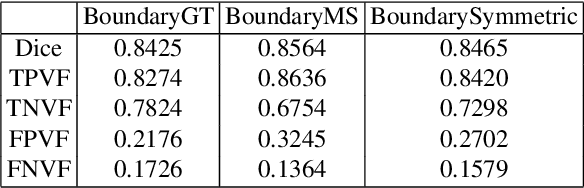
Abstract:Segmentation algorithms of medical image volumes are widely studied for many clinical and research purposes. We propose a novel and efficient framework for medical image segmentation. The framework functions under a deep learning paradigm, incorporating four novel contributions. Firstly, a residual interconnection is explored in different scale encoders. Secondly, four copy and crop connections are replaced to residual-block-based concatenation to alleviate the disparity between encoders and decoders, respectively. Thirdly, convolutional attention modules for feature refinement are studied on all scale decoders. Finally, an adaptive clean noisy label learning strategy(ACNLL) based on the training process from underfitting to overfitting is studied. Experimental results are illustrated on a publicly available benchmark database of spine CTs. Our segmentation framework achieves competitive performance with other state-of-the-art methods over a variety of different evaluation measures.
Deep Learning in Medical Ultrasound Image Segmentation: a Review
Feb 25, 2020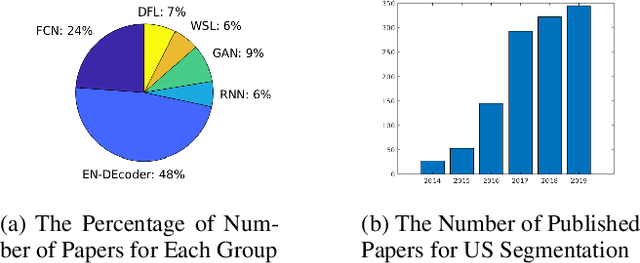
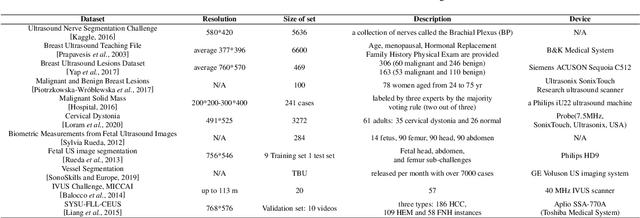
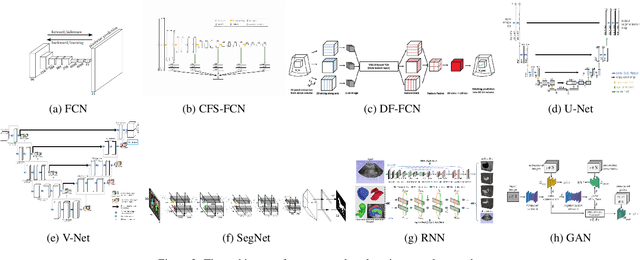
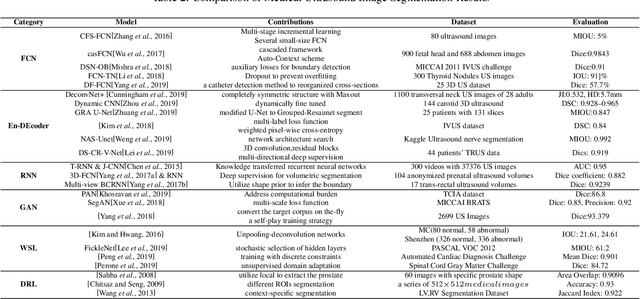
Abstract:Applying machine learning technologies, especially deep learning, into medical image segmentation is being widely studied because of its state-of-the-art performance and results. It can be a key step to provide a reliable basis for clinical diagnosis, such as 3D reconstruction of human tissues, image-guided interventions, image analyzing and visualization. In this review article, deep-learning-based methods for ultrasound image segmentation are categorized into six main groups according to their architectures and training at first. Secondly, for each group, several current representative algorithms are selected, introduced, analyzed and summarized in detail. In addition, common evaluation methods for image segmentation and ultrasound image segmentation datasets are summarized. Further, the performance of the current methods and their evaluations are reviewed. In the end, the challenges and potential research directions for medical ultrasound image segmentation are discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge