Huihua Yang
Boosting Generic Semi-Supervised Medical Image Segmentation via Diverse Teaching and Label Propagation
Aug 12, 2025Abstract:Both limited annotation and domain shift are significant challenges frequently encountered in medical image segmentation, leading to derivative scenarios like semi-supervised medical (SSMIS), semi-supervised medical domain generalization (Semi-MDG) and unsupervised medical domain adaptation (UMDA). Conventional methods are generally tailored to specific tasks in isolation, the error accumulation hinders the effective utilization of unlabeled data and limits further improvements, resulting in suboptimal performance when these issues occur. In this paper, we aim to develop a generic framework that masters all three tasks. We found that the key to solving the problem lies in how to generate reliable pseudo labels for the unlabeled data in the presence of domain shift with labeled data and increasing the diversity of the model. To tackle this issue, we employ a Diverse Teaching and Label Propagation Network (DTLP-Net) to boosting the Generic Semi-Supervised Medical Image Segmentation. Our DTLP-Net involves a single student model and two diverse teacher models, which can generate reliable pseudo-labels for the student model. The first teacher model decouple the training process with labeled and unlabeled data, The second teacher is momentum-updated periodically, thus generating reliable yet divers pseudo-labels. To fully utilize the information within the data, we adopt inter-sample and intra-sample data augmentation to learn the global and local knowledge. In addition, to further capture the voxel-level correlations, we propose label propagation to enhance the model robust. We evaluate our proposed framework on five benchmark datasets for SSMIS, UMDA, and Semi-MDG tasks. The results showcase notable improvements compared to state-of-the-art methods across all five settings, indicating the potential of our framework to tackle more challenging SSL scenarios.
UDCR: Unsupervised Aortic DSA/CTA Rigid Registration Using Deep Reinforcement Learning and Overlap Degree Calculation
Mar 09, 2024



Abstract:The rigid registration of aortic Digital Subtraction Angiography (DSA) and Computed Tomography Angiography (CTA) can provide 3D anatomical details of the vasculature for the interventional surgical treatment of conditions such as aortic dissection and aortic aneurysms, holding significant value for clinical research. However, the current methods for 2D/3D image registration are dependent on manual annotations or synthetic data, as well as the extraction of landmarks, which is not suitable for cross-modal registration of aortic DSA/CTA. In this paper, we propose an unsupervised method, UDCR, for aortic DSA/CTA rigid registration based on deep reinforcement learning. Leveraging the imaging principles and characteristics of DSA and CTA, we have constructed a cross-dimensional registration environment based on spatial transformations. Specifically, we propose an overlap degree calculation reward function that measures the intensity difference between the foreground and background, aimed at assessing the accuracy of registration between segmentation maps and DSA images. This method is highly flexible, allowing for the loading of pre-trained models to perform registration directly or to seek the optimal spatial transformation parameters through online learning. We manually annotated 61 pairs of aortic DSA/CTA for algorithm evaluation. The results indicate that the proposed UDCR achieved a Mean Absolute Error (MAE) of 2.85 mm in translation and 4.35{\deg} in rotation, showing significant potential for clinical applications.
Image-Guided Autonomous Guidewire Navigation in Robot-Assisted Endovascular Interventions using Reinforcement Learning
Mar 09, 2024



Abstract:Autonomous robots in endovascular interventions possess the potential to navigate guidewires with safety and reliability, while reducing human error and shortening surgical time. However, current methods of guidewire navigation based on Reinforcement Learning (RL) depend on manual demonstration data or magnetic guidance. In this work, we propose an Image-guided Autonomous Guidewire Navigation (IAGN) method. Specifically, we introduce BDA-star, a path planning algorithm with boundary distance constraints, for the trajectory planning of guidewire navigation. We established an IAGN-RL environment where the observations are real-time guidewire feeding images highlighting the position of the guidewire tip and the planned path. We proposed a reward function based on the distances from both the guidewire tip to the planned path and the target to evaluate the agent's actions. Furthermore, in policy network, we employ a pre-trained convolutional neural network to extract features, mitigating stability issues and slow convergence rates associated with direct learning from raw pixels. Experiments conducted on the aortic simulation IAGN platform demonstrated that the proposed method, targeting the left subclavian artery and the brachiocephalic artery, achieved a 100% guidewire navigation success rate, along with reduced movement and retraction distances and trajectories tend to the center of the vessels.
An Automatic Cascaded Model for Hemorrhagic Stroke Segmentation and Hemorrhagic Volume Estimation
Jan 09, 2024Abstract:Hemorrhagic Stroke (HS) has a rapid onset and is a serious condition that poses a great health threat. Promptly and accurately delineating the bleeding region and estimating the volume of bleeding in Computer Tomography (CT) images can assist clinicians in treatment planning, leading to improved treatment outcomes for patients. In this paper, a cascaded 3D model is constructed based on UNet to perform a two-stage segmentation of the hemorrhage area in CT images from rough to fine, and the hemorrhage volume is automatically calculated from the segmented area. On a dataset with 341 cases of hemorrhagic stroke CT scans, the proposed model provides high-quality segmentation outcome with higher accuracy (DSC 85.66%) and better computation efficiency (6.2 second per sample) when compared to the traditional Tada formula with respect to hemorrhage volume estimation.
Cross-head mutual Mean-Teaching for semi-supervised medical image segmentation
Oct 16, 2023Abstract:Semi-supervised medical image segmentation (SSMIS) has witnessed substantial advancements by leveraging limited labeled data and abundant unlabeled data. Nevertheless, existing state-of-the-art (SOTA) methods encounter challenges in accurately predicting labels for the unlabeled data, giving rise to disruptive noise during training and susceptibility to erroneous information overfitting. Moreover, applying perturbations to inaccurate predictions further reduces consistent learning. To address these concerns, we propose a novel Cross-head mutual mean-teaching Network (CMMT-Net) incorporated strong-weak data augmentation, thereby benefitting both self-training and consistency learning. Specifically, our CMMT-Net consists of both teacher-student peer networks with a share encoder and dual slightly different decoders, and the pseudo labels generated by one mean teacher head are adopted to supervise the other student branch to achieve a mutual consistency. Furthermore, we propose mutual virtual adversarial training (MVAT) to smooth the decision boundary and enhance feature representations. To diversify the consistency training samples, we employ Cross-Set CutMix strategy, which also helps address distribution mismatch issues. Notably, CMMT-Net simultaneously implements data, feature, and network perturbations, amplifying model diversity and generalization performance. Experimental results on three publicly available datasets indicate that our approach yields remarkable improvements over previous SOTA methods across various semi-supervised scenarios. Code and logs will be available at https://github.com/Leesoon1984/CMMT-Net.
Two-Stage Hybrid Supervision Framework for Fast, Low-resource, and Accurate Organ and Pan-cancer Segmentation in Abdomen CT
Sep 11, 2023



Abstract:Abdominal organ and tumour segmentation has many important clinical applications, such as organ quantification, surgical planning, and disease diagnosis. However, manual assessment is inherently subjective with considerable inter- and intra-expert variability. In the paper, we propose a hybrid supervised framework, StMt, that integrates self-training and mean teacher for the segmentation of abdominal organs and tumors using partially labeled and unlabeled data. We introduce a two-stage segmentation pipeline and whole-volume-based input strategy to maximize segmentation accuracy while meeting the requirements of inference time and GPU memory usage. Experiments on the validation set of FLARE2023 demonstrate that our method achieves excellent segmentation performance as well as fast and low-resource model inference. Our method achieved an average DSC score of 89.79\% and 45.55 \% for the organs and lesions on the validation set and the average running time and area under GPU memory-time cure are 11.25s and 9627.82MB, respectively.
TSI-Net: A Timing Sequence Image Segmentation Network for Intracranial Artery Segmentation in Digital Subtraction Angiography
Sep 07, 2023



Abstract:Cerebrovascular disease is one of the major diseases facing the world today. Automatic segmentation of intracranial artery (IA) in digital subtraction angiography (DSA) sequences is an important step in the diagnosis of vascular related diseases and in guiding neurointerventional procedures. While, a single image can only show part of the IA within the contrast medium according to the imaging principle of DSA technology. Therefore, 2D DSA segmentation methods are unable to capture the complete IA information and treatment of cerebrovascular diseases. We propose A timing sequence image segmentation network with U-shape, called TSI-Net, which incorporates a bi-directional ConvGRU module (BCM) in the encoder. The network incorporates a bi-directional ConvGRU module (BCM) in the encoder, which can input variable-length DSA sequences, retain past and future information, segment them into 2D images. In addition, we introduce a sensitive detail branch (SDB) at the end for supervising fine vessels. Experimented on the DSA sequence dataset DIAS, the method performs significantly better than state-of-the-art networks in recent years. In particular, it achieves a Sen evaluation metric of 0.797, which is a 3% improvement compared to other methods.
DIAS: A Comprehensive Benchmark for DSA-sequence Intracranial Artery Segmentation
Jun 21, 2023



Abstract:Automatic segmentation of the intracranial artery (IA) in digital subtraction angiography (DSA) sequence is an essential step in diagnosing IA-related diseases and guiding neuro-interventional surgery. However, the lack of publicly available datasets has impeded research in this area. In this paper, we release DIAS, an IA segmentation dataset, consisting of 120 DSA sequences from intracranial interventional therapy. In addition to pixel-wise annotations, this dataset provides two types of scribble annotations for weakly supervised IA segmentation research. We present a comprehensive benchmark for evaluating the performance of this challenging dataset by utilizing fully-, weakly-, and semi-supervised learning approaches. Specifically, we propose a method that incorporates a dimensionality reduction module into a 2D/3D model to achieve vessel segmentation in DSA sequences. For weakly-supervised learning, we propose a scribble learning-based image segmentation framework, SSCR, which comprises scribble supervision and consistency regularization. Furthermore, we introduce a random patch-based self-training framework that utilizes unlabeled DSA sequences to improve segmentation performance. Our extensive experiments on the DIAS dataset demonstrate the effectiveness of these methods as potential baselines for future research and clinical applications.
Combining Hybrid Architecture and Pseudo-label for Semi-supervised Abdominal Organ Segmentation
Jul 23, 2022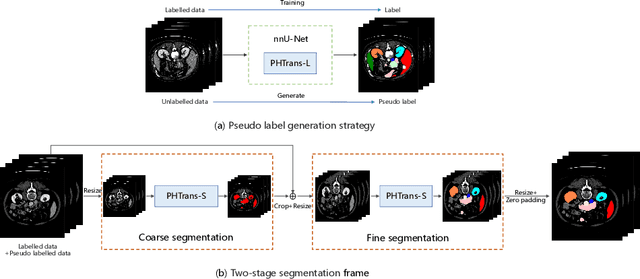

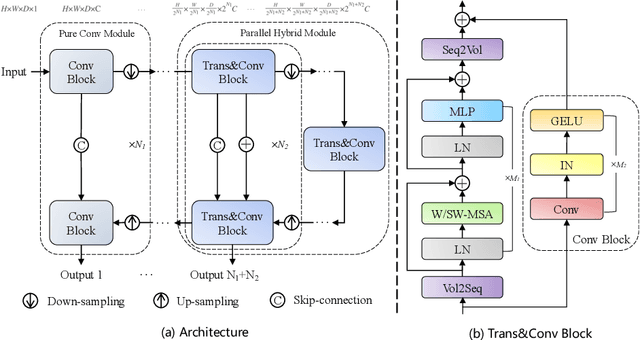
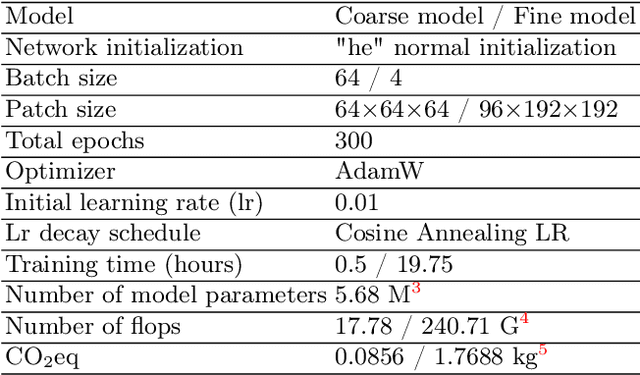
Abstract:Abdominal organ segmentation has many important clinical applications, such as organ quantification, surgical planning, and disease diagnosis. However, manually annotating organs from CT scans is time-consuming and labor-intensive. Semi-supervised learning has shown the potential to alleviate this challenge by learning from a large set of unlabeled images and limited labeled samples. In this work, we follow the self-training strategy and employ a hybrid architecture (PHTrans) with CNN and Transformer for both teacher and student models to generate precise pseudo-labels. Afterward, we introduce them with label data together into a two-stage segmentation framework with lightweight PHTrans for training to improve the performance and generalization ability of the model while remaining efficient. Experiments on the validation set of FLARE2022 demonstrate that our method achieves excellent segmentation performance as well as fast and low-resource model inference. The average DSC and HSD are 0.8956 and 0.9316, respectively. Under our development environments, the average inference time is 18.62 s, the average maximum GPU memory is 1995.04 MB, and the area under the GPU memory-time curve and the average area under the CPU utilization-time curve are 23196.84 and 319.67.
WSSS4LUAD: Grand Challenge on Weakly-supervised Tissue Semantic Segmentation for Lung Adenocarcinoma
Apr 14, 2022
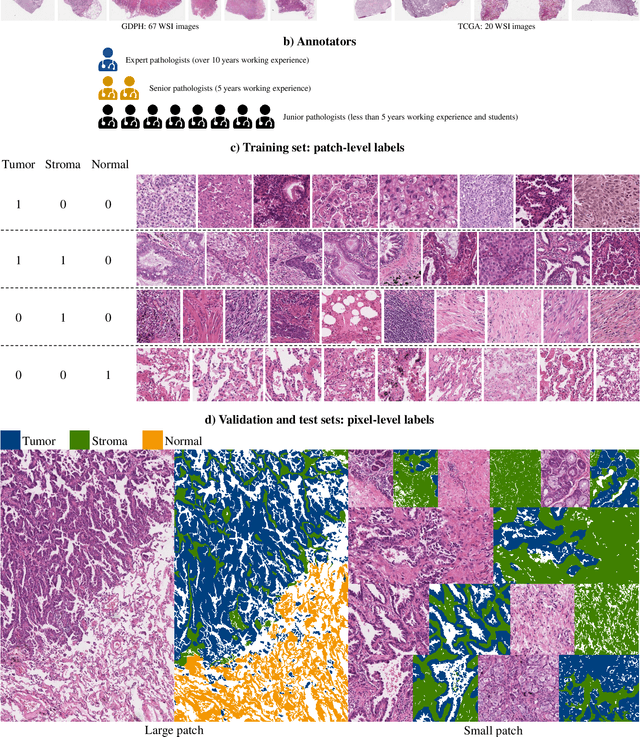
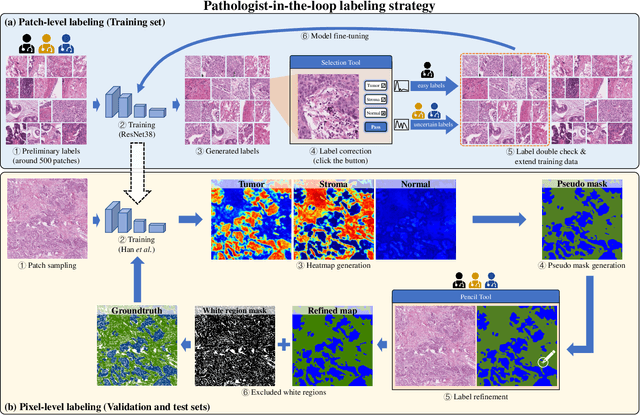
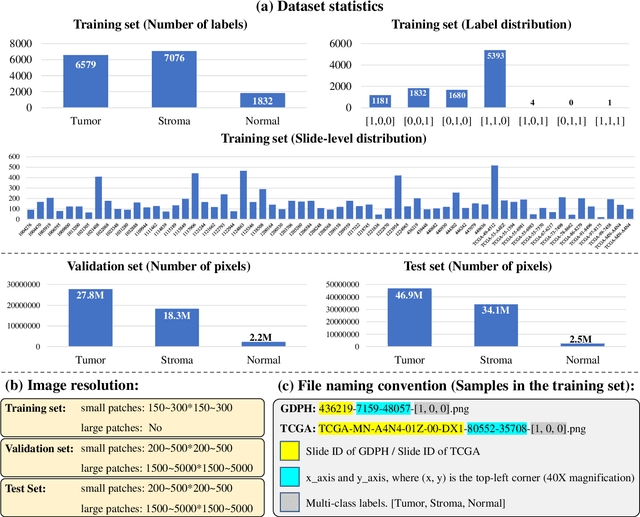
Abstract:Lung cancer is the leading cause of cancer death worldwide, and adenocarcinoma (LUAD) is the most common subtype. Exploiting the potential value of the histopathology images can promote precision medicine in oncology. Tissue segmentation is the basic upstream task of histopathology image analysis. Existing deep learning models have achieved superior segmentation performance but require sufficient pixel-level annotations, which is time-consuming and expensive. To enrich the label resources of LUAD and to alleviate the annotation efforts, we organize this challenge WSSS4LUAD to call for the outstanding weakly-supervised semantic segmentation (WSSS) techniques for histopathology images of LUAD. Participants have to design the algorithm to segment tumor epithelial, tumor-associated stroma and normal tissue with only patch-level labels. This challenge includes 10,091 patch-level annotations (the training set) and over 130 million labeled pixels (the validation and test sets), from 87 WSIs (67 from GDPH, 20 from TCGA). All the labels were generated by a pathologist-in-the-loop pipeline with the help of AI models and checked by the label review board. Among 532 registrations, 28 teams submitted the results in the test phase with over 1,000 submissions. Finally, the first place team achieved mIoU of 0.8413 (tumor: 0.8389, stroma: 0.7931, normal: 0.8919). According to the technical reports of the top-tier teams, CAM is still the most popular approach in WSSS. Cutmix data augmentation has been widely adopted to generate more reliable samples. With the success of this challenge, we believe that WSSS approaches with patch-level annotations can be a complement to the traditional pixel annotations while reducing the annotation efforts. The entire dataset has been released to encourage more researches on computational pathology in LUAD and more novel WSSS techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge