Guanyu Yang
LTSI, CRIBS, LIST
Diff-OP3D: Bridging 2D Diffusion for Open Pose 3D Zero-Shot Classification
Dec 12, 2023Abstract:With the explosive 3D data growth, the urgency of utilizing zero-shot learning to facilitate data labeling becomes evident. Recently, the methods via transferring Contrastive Language-Image Pre-training (CLIP) to 3D vision have made great progress in the 3D zero-shot classification task. However, these methods primarily focus on aligned pose 3D objects (ap-3os), overlooking the recognition of 3D objects with open poses (op-3os) typically encountered in real-world scenarios, such as an overturned chair or a lying teddy bear. To this end, we propose a more challenging benchmark for 3D open-pose zero-shot classification. Echoing our benchmark, we design a concise angle-refinement mechanism that automatically optimizes one ideal pose as well as classifies these op-3os. Furthermore, we make a first attempt to bridge 2D pre-trained diffusion model as a classifer to 3D zero-shot classification without any additional training. Such 2D diffusion to 3D objects proves vital in improving zero-shot classification for both ap-3os and op-3os. Our model notably improves by 3.5% and 15.8% on ModelNet10$^{\ddag}$ and McGill$^{\ddag}$ open pose benchmarks, respectively, and surpasses the current state-of-the-art by 6.8% on the aligned pose ModelNet10, affirming diffusion's efficacy in 3D zero-shot tasks.
Knowledge Boosting: Rethinking Medical Contrastive Vision-Language Pre-Training
Jul 17, 2023



Abstract:The foundation models based on pre-training technology have significantly advanced artificial intelligence from theoretical to practical applications. These models have facilitated the feasibility of computer-aided diagnosis for widespread use. Medical contrastive vision-language pre-training, which does not require human annotations, is an effective approach for guiding representation learning using description information in diagnostic reports. However, the effectiveness of pre-training is limited by the large-scale semantic overlap and shifting problems in medical field. To address these issues, we propose the Knowledge-Boosting Contrastive Vision-Language Pre-training framework (KoBo), which integrates clinical knowledge into the learning of vision-language semantic consistency. The framework uses an unbiased, open-set sample-wise knowledge representation to measure negative sample noise and supplement the correspondence between vision-language mutual information and clinical knowledge. Extensive experiments validate the effect of our framework on eight tasks including classification, segmentation, retrieval, and semantic relatedness, achieving comparable or better performance with the zero-shot or few-shot settings. Our code is open on https://github.com/ChenXiaoFei-CS/KoBo.
Dynamic Snake Convolution based on Topological Geometric Constraints for Tubular Structure Segmentation
Jul 17, 2023



Abstract:Accurate segmentation of topological tubular structures, such as blood vessels and roads, is crucial in various fields, ensuring accuracy and efficiency in downstream tasks. However, many factors complicate the task, including thin local structures and variable global morphologies. In this work, we note the specificity of tubular structures and use this knowledge to guide our DSCNet to simultaneously enhance perception in three stages: feature extraction, feature fusion, and loss constraint. First, we propose a dynamic snake convolution to accurately capture the features of tubular structures by adaptively focusing on slender and tortuous local structures. Subsequently, we propose a multi-view feature fusion strategy to complement the attention to features from multiple perspectives during feature fusion, ensuring the retention of important information from different global morphologies. Finally, a continuity constraint loss function, based on persistent homology, is proposed to constrain the topological continuity of the segmentation better. Experiments on 2D and 3D datasets show that our DSCNet provides better accuracy and continuity on the tubular structure segmentation task compared with several methods. Our codes will be publicly available.
Partial Vessels Annotation-based Coronary Artery Segmentation with Self-training and Prototype Learning
Jul 10, 2023



Abstract:Coronary artery segmentation on coronary-computed tomography angiography (CCTA) images is crucial for clinical use. Due to the expertise-required and labor-intensive annotation process, there is a growing demand for the relevant label-efficient learning algorithms. To this end, we propose partial vessels annotation (PVA) based on the challenges of coronary artery segmentation and clinical diagnostic characteristics. Further, we propose a progressive weakly supervised learning framework to achieve accurate segmentation under PVA. First, our proposed framework learns the local features of vessels to propagate the knowledge to unlabeled regions. Subsequently, it learns the global structure by utilizing the propagated knowledge, and corrects the errors introduced in the propagation process. Finally, it leverages the similarity between feature embeddings and the feature prototype to enhance testing outputs. Experiments on clinical data reveals that our proposed framework outperforms the competing methods under PVA (24.29% vessels), and achieves comparable performance in trunk continuity with the baseline model using full annotation (100% vessels).
Geometric Visual Similarity Learning in 3D Medical Image Self-supervised Pre-training
Mar 02, 2023Abstract:Learning inter-image similarity is crucial for 3D medical images self-supervised pre-training, due to their sharing of numerous same semantic regions. However, the lack of the semantic prior in metrics and the semantic-independent variation in 3D medical images make it challenging to get a reliable measurement for the inter-image similarity, hindering the learning of consistent representation for same semantics. We investigate the challenging problem of this task, i.e., learning a consistent representation between images for a clustering effect of same semantic features. We propose a novel visual similarity learning paradigm, Geometric Visual Similarity Learning, which embeds the prior of topological invariance into the measurement of the inter-image similarity for consistent representation of semantic regions. To drive this paradigm, we further construct a novel geometric matching head, the Z-matching head, to collaboratively learn the global and local similarity of semantic regions, guiding the efficient representation learning for different scale-level inter-image semantic features. Our experiments demonstrate that the pre-training with our learning of inter-image similarity yields more powerful inner-scene, inter-scene, and global-local transferring ability on four challenging 3D medical image tasks. Our codes and pre-trained models will be publicly available on https://github.com/YutingHe-list/GVSL.
* Accepted by CVPR 2023
Rebalanced Zero-shot Learning
Oct 13, 2022
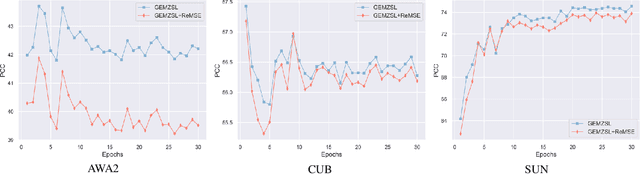
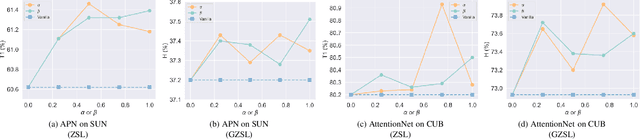

Abstract:Zero-shot learning (ZSL) aims to identify unseen classes with zero samples during training. Broadly speaking, present ZSL methods usually adopt class-level semantic labels and compare them with instance-level semantic predictions to infer unseen classes. However, we find that such existing models mostly produce imbalanced semantic predictions, i.e. these models could perform precisely for some semantics, but may not for others. To address the drawback, we aim to introduce an imbalanced learning framework into ZSL. However, we find that imbalanced ZSL has two unique challenges: (1) Its imbalanced predictions are highly correlated with the value of semantic labels rather than the number of samples as typically considered in the traditional imbalanced learning; (2) Different semantics follow quite different error distributions between classes. To mitigate these issues, we first formalize ZSL as an imbalanced regression problem which offers theoretical foundations to interpret how semantic labels lead to imbalanced semantic predictions. We then propose a re-weighted loss termed Re-balanced Mean-Squared Error (ReMSE), which tracks the mean and variance of error distributions, thus ensuring rebalanced learning across classes. As a major contribution, we conduct a series of analyses showing that ReMSE is theoretically well established. Extensive experiments demonstrate that the proposed method effectively alleviates the imbalance in semantic prediction and outperforms many state-of-the-art ZSL methods.
FFCNet: Fourier Transform-Based Frequency Learning and Complex Convolutional Network for Colon Disease Classification
Jul 04, 2022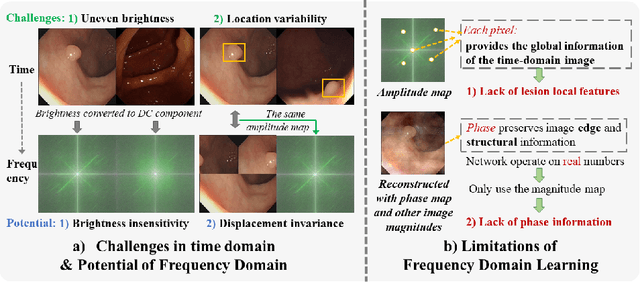
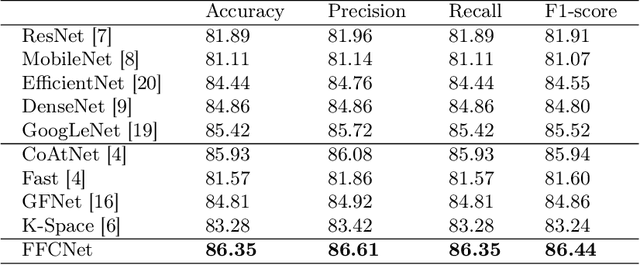
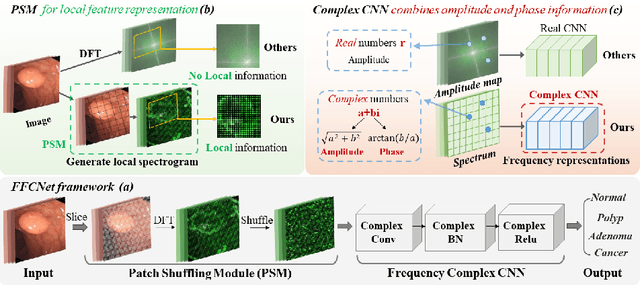
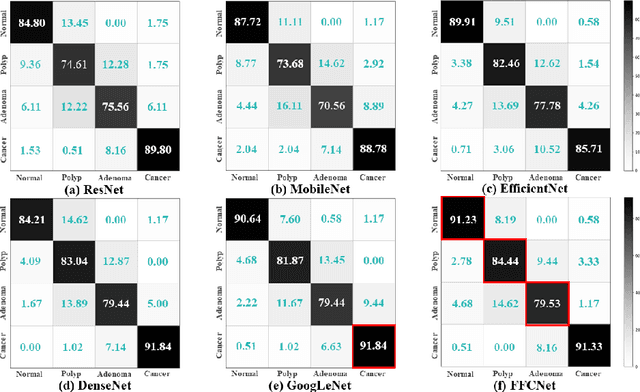
Abstract:Reliable automatic classification of colonoscopy images is of great significance in assessing the stage of colonic lesions and formulating appropriate treatment plans. However, it is challenging due to uneven brightness, location variability, inter-class similarity, and intra-class dissimilarity, affecting the classification accuracy. To address the above issues, we propose a Fourier-based Frequency Complex Network (FFCNet) for colon disease classification in this study. Specifically, FFCNet is a novel complex network that enables the combination of complex convolutional networks with frequency learning to overcome the loss of phase information caused by real convolution operations. Also, our Fourier transform transfers the average brightness of an image to a point in the spectrum (the DC component), alleviating the effects of uneven brightness by decoupling image content and brightness. Moreover, the image patch scrambling module in FFCNet generates random local spectral blocks, empowering the network to learn long-range and local diseasespecific features and improving the discriminative ability of hard samples. We evaluated the proposed FFCNet on an in-house dataset with 2568 colonoscopy images, showing our method achieves high performance outperforming previous state-of-the art methods with an accuracy of 86:35% and an accuracy of 4.46% higher than the backbone. The project page with code is available at https://github.com/soleilssss/FFCNet.
Dual Contrastive Attributed Graph Clustering Network
Jun 16, 2022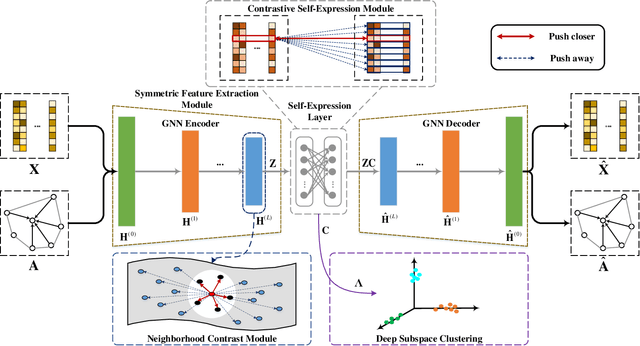
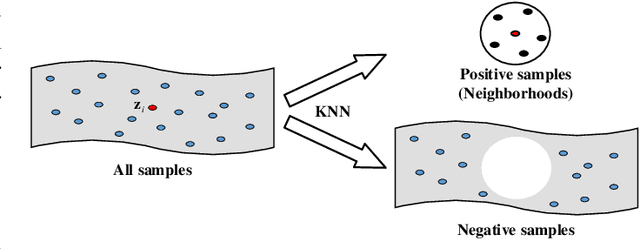
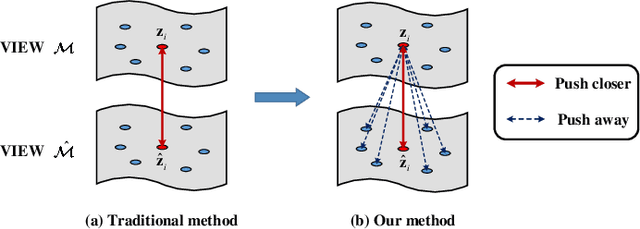
Abstract:Attributed graph clustering is one of the most important tasks in graph analysis field, the goal of which is to group nodes with similar representations into the same cluster without manual guidance. Recent studies based on graph contrastive learning have achieved impressive results in processing graph-structured data. However, existing graph contrastive learning based methods 1) do not directly address the clustering task, since the representation learning and clustering process are separated; 2) depend too much on graph data augmentation, which greatly limits the capability of contrastive learning; 3) ignore the contrastive message for subspace clustering. To accommodate the aforementioned issues, we propose a generic framework called Dual Contrastive Attributed Graph Clustering Network (DCAGC). In DCAGC, by leveraging Neighborhood Contrast Module, the similarity of the neighbor nodes will be maximized and the quality of the node representation will be improved. Meanwhile, the Contrastive Self-Expression Module is built by minimizing the node representation before and after the reconstruction of the self-expression layer to obtain a discriminative self-expression matrix for spectral clustering. All the modules of DCAGC are trained and optimized in a unified framework, so the learned node representation contains clustering-oriented messages. Extensive experimental results on four attributed graph datasets show the superiority of DCAGC compared with 16 state-of-the-art clustering methods. The code of this paper is available at https://github.com/wangtong627/Dual-Contrastive-Attributed-Graph-Clustering-Network.
XMorpher: Full Transformer for Deformable Medical Image Registration via Cross Attention
Jun 15, 2022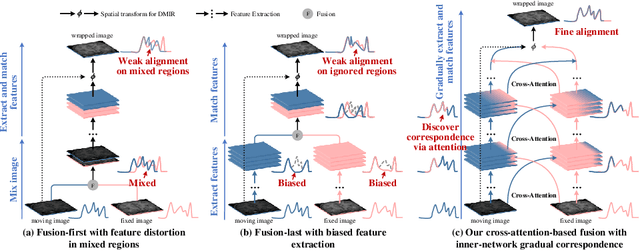
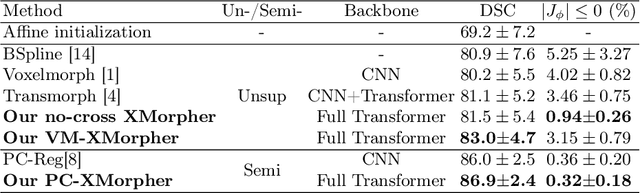
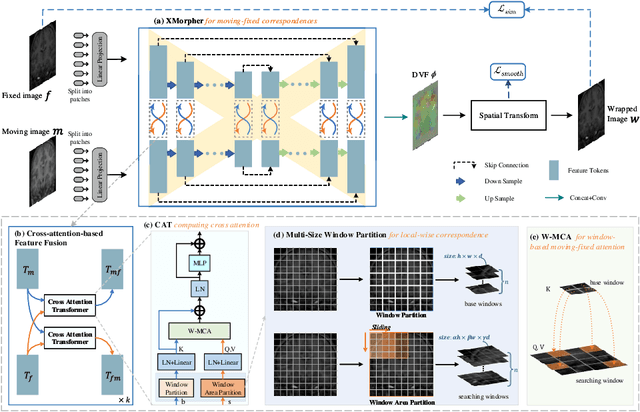
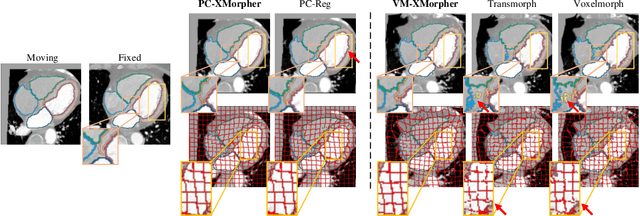
Abstract:An effective backbone network is important to deep learning-based Deformable Medical Image Registration (DMIR), because it extracts and matches the features between two images to discover the mutual correspondence for fine registration. However, the existing deep networks focus on single image situation and are limited in registration task which is performed on paired images. Therefore, we advance a novel backbone network, XMorpher, for the effective corresponding feature representation in DMIR. 1) It proposes a novel full transformer architecture including dual parallel feature extraction networks which exchange information through cross attention, thus discovering multi-level semantic correspondence while extracting respective features gradually for final effective registration. 2) It advances the Cross Attention Transformer (CAT) blocks to establish the attention mechanism between images which is able to find the correspondence automatically and prompts the features to fuse efficiently in the network. 3) It constrains the attention computation between base windows and searching windows with different sizes, and thus focuses on the local transformation of deformable registration and enhances the computing efficiency at the same time. Without any bells and whistles, our XMorpher gives Voxelmorph 2.8% improvement on DSC , demonstrating its effective representation of the features from the paired images in DMIR. We believe that our XMorpher has great application potential in more paired medical images. Our XMorpher is open on https://github.com/Solemoon/XMorpher
MNet: Rethinking 2D/3D Networks for Anisotropic Medical Image Segmentation
May 10, 2022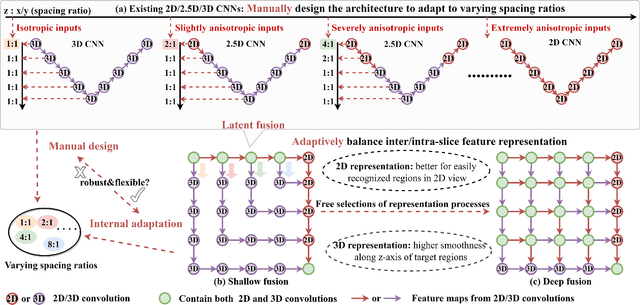
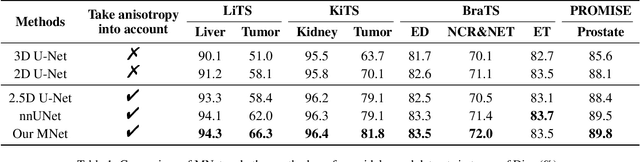
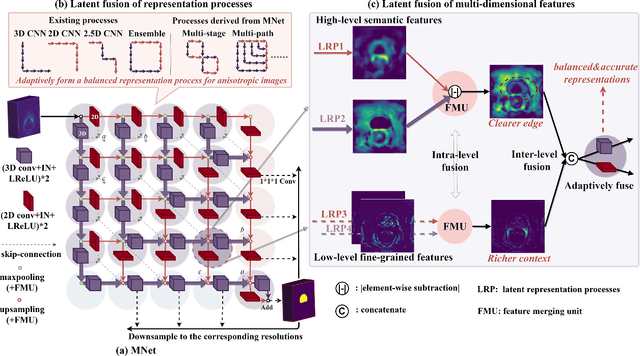

Abstract:The nature of thick-slice scanning causes severe inter-slice discontinuities of 3D medical images, and the vanilla 2D/3D convolutional neural networks (CNNs) fail to represent sparse inter-slice information and dense intra-slice information in a balanced way, leading to severe underfitting to inter-slice features (for vanilla 2D CNNs) and overfitting to noise from long-range slices (for vanilla 3D CNNs). In this work, a novel mesh network (MNet) is proposed to balance the spatial representation inter axes via learning. 1) Our MNet latently fuses plenty of representation processes by embedding multi-dimensional convolutions deeply into basic modules, making the selections of representation processes flexible, thus balancing representation for sparse inter-slice information and dense intra-slice information adaptively. 2) Our MNet latently fuses multi-dimensional features inside each basic module, simultaneously taking the advantages of 2D (high segmentation accuracy of the easily recognized regions in 2D view) and 3D (high smoothness of 3D organ contour) representations, thus obtaining more accurate modeling for target regions. Comprehensive experiments are performed on four public datasets (CT\&MR), the results consistently demonstrate the proposed MNet outperforms the other methods. The code and datasets are available at: https://github.com/zfdong-code/MNet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge