David Rolnick
Mila Quebec AI Institute, Canada, McGill University, Canada
Alberta Wells Dataset: Pinpointing Oil and Gas Wells from Satellite Imagery
Oct 11, 2024



Abstract:Millions of abandoned oil and gas wells are scattered across the world, leaching methane into the atmosphere and toxic compounds into the groundwater. Many of these locations are unknown, preventing the wells from being plugged and their polluting effects averted. Remote sensing is a relatively unexplored tool for pinpointing abandoned wells at scale. We introduce the first large-scale benchmark dataset for this problem, leveraging medium-resolution multi-spectral satellite imagery from Planet Labs. Our curated dataset comprises over 213,000 wells (abandoned, suspended, and active) from Alberta, a region with especially high well density, sourced from the Alberta Energy Regulator and verified by domain experts. We evaluate baseline algorithms for well detection and segmentation, showing the promise of computer vision approaches but also significant room for improvement.
Causal Representation Learning in Temporal Data via Single-Parent Decoding
Oct 09, 2024



Abstract:Scientific research often seeks to understand the causal structure underlying high-level variables in a system. For example, climate scientists study how phenomena, such as El Ni\~no, affect other climate processes at remote locations across the globe. However, scientists typically collect low-level measurements, such as geographically distributed temperature readings. From these, one needs to learn both a mapping to causally-relevant latent variables, such as a high-level representation of the El Ni\~no phenomenon and other processes, as well as the causal model over them. The challenge is that this task, called causal representation learning, is highly underdetermined from observational data alone, requiring other constraints during learning to resolve the indeterminacies. In this work, we consider a temporal model with a sparsity assumption, namely single-parent decoding: each observed low-level variable is only affected by a single latent variable. Such an assumption is reasonable in many scientific applications that require finding groups of low-level variables, such as extracting regions from geographically gridded measurement data in climate research or capturing brain regions from neural activity data. We demonstrate the identifiability of the resulting model and propose a differentiable method, Causal Discovery with Single-parent Decoding (CDSD), that simultaneously learns the underlying latents and a causal graph over them. We assess the validity of our theoretical results using simulated data and showcase the practical validity of our method in an application to real-world data from the climate science field.
Tree semantic segmentation from aerial image time series
Jul 18, 2024



Abstract:Earth's forests play an important role in the fight against climate change, and are in turn negatively affected by it. Effective monitoring of different tree species is essential to understanding and improving the health and biodiversity of forests. In this work, we address the challenge of tree species identification by performing semantic segmentation of trees using an aerial image dataset spanning over a year. We compare models trained on single images versus those trained on time series to assess the impact of tree phenology on segmentation performances. We also introduce a simple convolutional block for extracting spatio-temporal features from image time series, enabling the use of popular pretrained backbones and methods. We leverage the hierarchical structure of tree species taxonomy by incorporating a custom loss function that refines predictions at three levels: species, genus, and higher-level taxa. Our findings demonstrate the superiority of our methodology in exploiting the time series modality and confirm that enriching labels using taxonomic information improves the semantic segmentation performance.
Evaluating the transferability potential of deep learning models for climate downscaling
Jul 17, 2024



Abstract:Climate downscaling, the process of generating high-resolution climate data from low-resolution simulations, is essential for understanding and adapting to climate change at regional and local scales. Deep learning approaches have proven useful in tackling this problem. However, existing studies usually focus on training models for one specific task, location and variable, which are therefore limited in their generalizability and transferability. In this paper, we evaluate the efficacy of training deep learning downscaling models on multiple diverse climate datasets to learn more robust and transferable representations. We evaluate the effectiveness of architectures zero-shot transferability using CNNs, Fourier Neural Operators (FNOs), and vision Transformers (ViTs). We assess the spatial, variable, and product transferability of downscaling models experimentally, to understand the generalizability of these different architecture types.
Improving Molecular Modeling with Geometric GNNs: an Empirical Study
Jul 11, 2024



Abstract:Rapid advancements in machine learning (ML) are transforming materials science by significantly speeding up material property calculations. However, the proliferation of ML approaches has made it challenging for scientists to keep up with the most promising techniques. This paper presents an empirical study on Geometric Graph Neural Networks for 3D atomic systems, focusing on the impact of different (1) canonicalization methods, (2) graph creation strategies, and (3) auxiliary tasks, on performance, scalability and symmetry enforcement. Our findings and insights aim to guide researchers in selecting optimal modeling components for molecular modeling tasks.
Insect Identification in the Wild: The AMI Dataset
Jun 18, 2024



Abstract:Insects represent half of all global biodiversity, yet many of the world's insects are disappearing, with severe implications for ecosystems and agriculture. Despite this crisis, data on insect diversity and abundance remain woefully inadequate, due to the scarcity of human experts and the lack of scalable tools for monitoring. Ecologists have started to adopt camera traps to record and study insects, and have proposed computer vision algorithms as an answer for scalable data processing. However, insect monitoring in the wild poses unique challenges that have not yet been addressed within computer vision, including the combination of long-tailed data, extremely similar classes, and significant distribution shifts. We provide the first large-scale machine learning benchmarks for fine-grained insect recognition, designed to match real-world tasks faced by ecologists. Our contributions include a curated dataset of images from citizen science platforms and museums, and an expert-annotated dataset drawn from automated camera traps across multiple continents, designed to test out-of-distribution generalization under field conditions. We train and evaluate a variety of baseline algorithms and introduce a combination of data augmentation techniques that enhance generalization across geographies and hardware setups. Code and datasets are made publicly available.
A machine learning pipeline for automated insect monitoring
Jun 18, 2024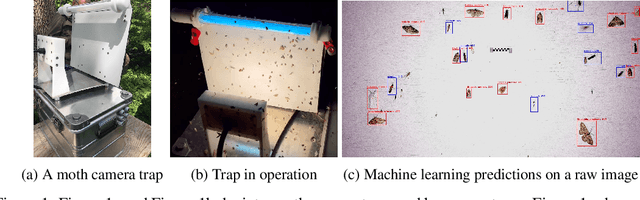
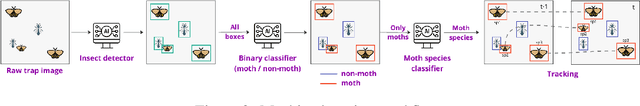
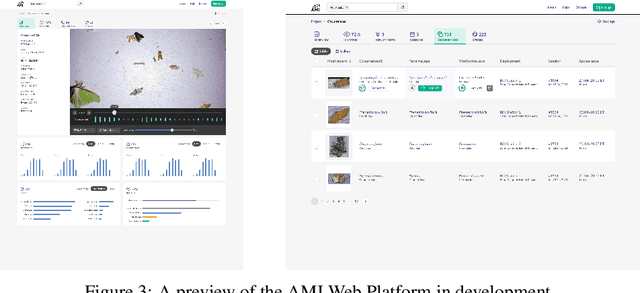

Abstract:Climate change and other anthropogenic factors have led to a catastrophic decline in insects, endangering both biodiversity and the ecosystem services on which human society depends. Data on insect abundance, however, remains woefully inadequate. Camera traps, conventionally used for monitoring terrestrial vertebrates, are now being modified for insects, especially moths. We describe a complete, open-source machine learning-based software pipeline for automated monitoring of moths via camera traps, including object detection, moth/non-moth classification, fine-grained identification of moth species, and tracking individuals. We believe that our tools, which are already in use across three continents, represent the future of massively scalable data collection in entomology.
Climate Variable Downscaling with Conditional Normalizing Flows
May 31, 2024



Abstract:Predictions of global climate models typically operate on coarse spatial scales due to the large computational costs of climate simulations. This has led to a considerable interest in methods for statistical downscaling, a similar process to super-resolution in the computer vision context, to provide more local and regional climate information. In this work, we apply conditional normalizing flows to the task of climate variable downscaling. We showcase its successful performance on an ERA5 water content dataset for different upsampling factors. Additionally, we show that the method allows us to assess the predictive uncertainty in terms of standard deviation from the fitted conditional distribution mean.
Simultaneous linear connectivity of neural networks modulo permutation
Apr 09, 2024Abstract:Neural networks typically exhibit permutation symmetries which contribute to the non-convexity of the networks' loss landscapes, since linearly interpolating between two permuted versions of a trained network tends to encounter a high loss barrier. Recent work has argued that permutation symmetries are the only sources of non-convexity, meaning there are essentially no such barriers between trained networks if they are permuted appropriately. In this work, we refine these arguments into three distinct claims of increasing strength. We show that existing evidence only supports "weak linear connectivity"-that for each pair of networks belonging to a set of SGD solutions, there exist (multiple) permutations that linearly connect it with the other networks. In contrast, the claim "strong linear connectivity"-that for each network, there exists one permutation that simultaneously connects it with the other networks-is both intuitively and practically more desirable. This stronger claim would imply that the loss landscape is convex after accounting for permutation, and enable linear interpolation between three or more independently trained models without increased loss. In this work, we introduce an intermediate claim-that for certain sequences of networks, there exists one permutation that simultaneously aligns matching pairs of networks from these sequences. Specifically, we discover that a single permutation aligns sequences of iteratively trained as well as iteratively pruned networks, meaning that two networks exhibit low loss barriers at each step of their optimization and sparsification trajectories respectively. Finally, we provide the first evidence that strong linear connectivity may be possible under certain conditions, by showing that barriers decrease with increasing network width when interpolating among three networks.
Predicting Species Occurrence Patterns from Partial Observations
Mar 28, 2024



Abstract:To address the interlinked biodiversity and climate crises, we need an understanding of where species occur and how these patterns are changing. However, observational data on most species remains very limited, and the amount of data available varies greatly between taxonomic groups. We introduce the problem of predicting species occurrence patterns given (a) satellite imagery, and (b) known information on the occurrence of other species. To evaluate algorithms on this task, we introduce SatButterfly, a dataset of satellite images, environmental data and observational data for butterflies, which is designed to pair with the existing SatBird dataset of bird observational data. To address this task, we propose a general model, R-Tran, for predicting species occurrence patterns that enables the use of partial observational data wherever found. We find that R-Tran outperforms other methods in predicting species encounter rates with partial information both within a taxon (birds) and across taxa (birds and butterflies). Our approach opens new perspectives to leveraging insights from species with abundant data to other species with scarce data, by modelling the ecosystems in which they co-occur.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge