Chia-Yen Lee
Predictability Analysis of Regression Problems via Conditional Entropy Estimations
Jun 06, 2024Abstract:In the field of machine learning, regression problems are pivotal due to their ability to predict continuous outcomes. Traditional error metrics like mean squared error, mean absolute error, and coefficient of determination measure model accuracy. The model accuracy is the consequence of the selected model and the features, which blurs the analysis of contribution. Predictability, in the other hand, focus on the predictable level of a target variable given a set of features. This study introduces conditional entropy estimators to assess predictability in regression problems, bridging this gap. We enhance and develop reliable conditional entropy estimators, particularly the KNIFE-P estimator and LMC-P estimator, which offer under- and over-estimation, providing a practical framework for predictability analysis. Extensive experiments on synthesized and real-world datasets demonstrate the robustness and utility of these estimators. Additionally, we extend the analysis to the coefficient of determination \(R^2 \), enhancing the interpretability of predictability. The results highlight the effectiveness of KNIFE-P and LMC-P in capturing the achievable performance and limitations of feature sets, providing valuable tools in the development of regression models. These indicators offer a robust framework for assessing the predictability for regression problems.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Airway Tree Modeling Using Dual-channel 3D UNet 3+ with Vesselness Prior
Aug 30, 2022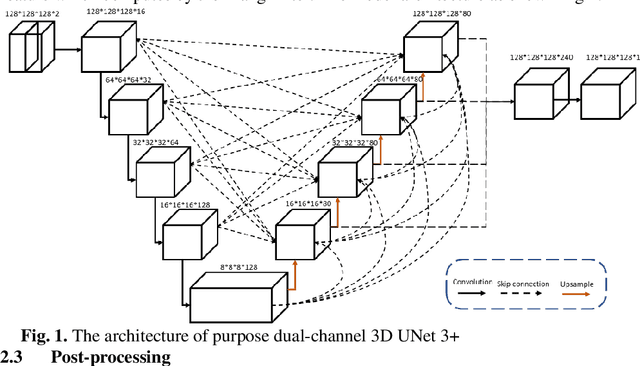
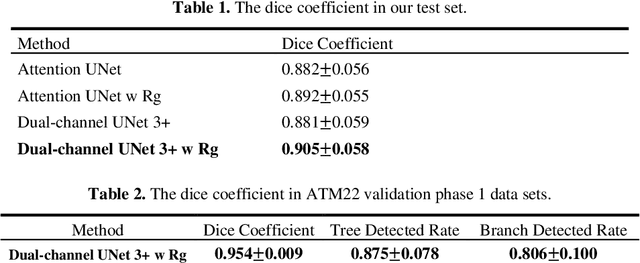
Abstract:The lung airway tree modeling is essential to work for the diagnosis of pulmonary diseases, especially for X-Ray computed tomography (CT). The airway tree modeling on CT images can provide the experts with 3-dimension measurements like wall thickness, etc. This information can tremendously aid the diagnosis of pulmonary diseases like chronic obstructive pulmonary disease [1-4]. Many scholars have attempted various ways to model the lung airway tree, which can be split into two major categories based on its nature. Namely, the model-based approach and the deep learning approach. The performance of a typical model-based approach usually depends on the manual tuning of the model parameter, which can be its advantages and disadvantages. The advantage is its don't require a large amount of training data which can be beneficial for a small dataset like medical imaging. On the other hand, the performance of model-based may be a misconcep-tion [5,6]. In recent years, deep learning has achieved good results in the field of medical image processing, and many scholars have used UNet-based methods in medical image segmentation [7-11]. Among all the variation of UNet, the UNet 3+ [11] have relatively good result compare to the rest of the variation of UNet. Therefor to further improve the accuracy of lung airway tree modeling, this study combines the Frangi filter [5] with UNet 3+ [11] to develop a dual-channel 3D UNet 3+. The Frangi filter is used to extracting vessel-like feature. The vessel-like feature then used as input to guide the dual-channel UNet 3+ training and testing procedures.
Using Multi-scale SwinTransformer-HTC with Data augmentation in CoNIC Challenge
Mar 02, 2022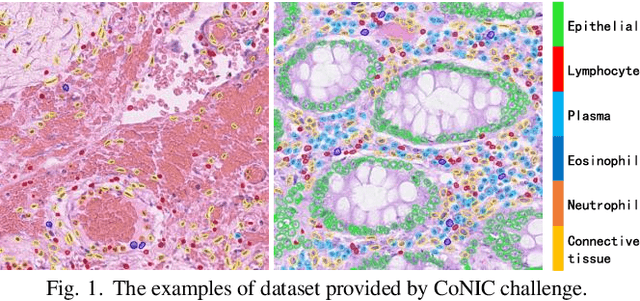
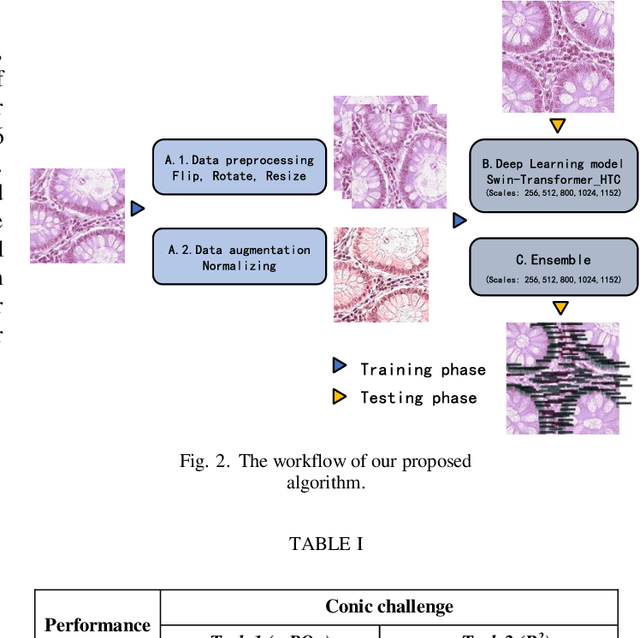
Abstract:Colorectal cancer is one of the most common cancers worldwide, so early pathological examination is very important. However, it is time-consuming and labor-intensive to identify the number and type of cells on H&E images in clinical. Therefore, automatic segmentation and classification task and counting the cellular composition of H&E images from pathological sections is proposed by CoNIC Challenge 2022. We proposed a multi-scale Swin transformer with HTC for this challenge, and also applied the known normalization methods to generate more augmentation data. Finally, our strategy showed that the multi-scale played a crucial role to identify different scale features and the augmentation arose the recognition of model.
ADAM Challenge: Detecting Age-related Macular Degeneration from Fundus Images
Feb 18, 2022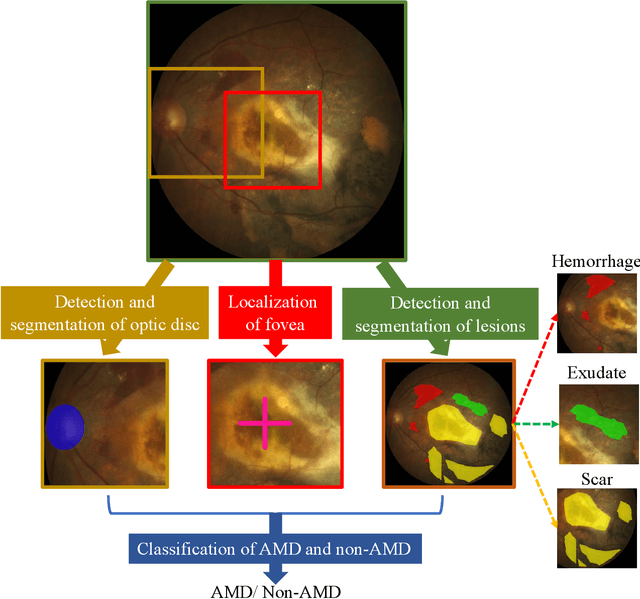
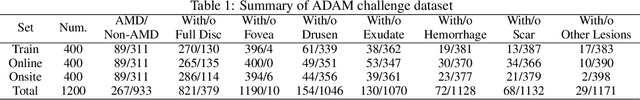
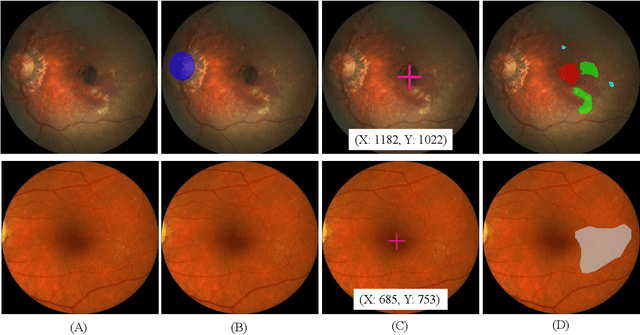
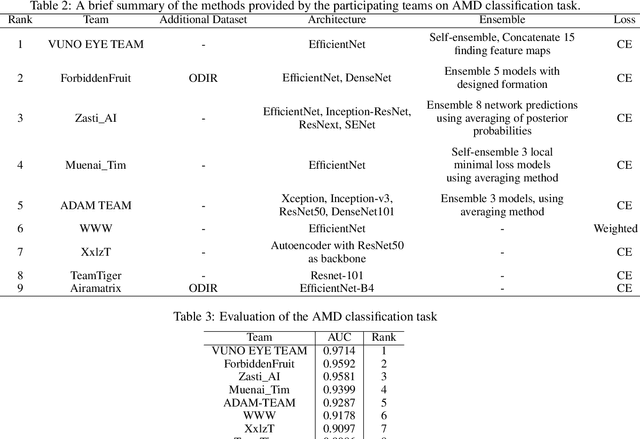
Abstract:Age-related macular degeneration (AMD) is the leading cause of visual impairment among elderly in the world. Early detection of AMD is of great importance as the vision loss caused by AMD is irreversible and permanent. Color fundus photography is the most cost-effective imaging modality to screen for retinal disorders. \textcolor{red}{Recently, some algorithms based on deep learning had been developed for fundus image analysis and automatic AMD detection. However, a comprehensive annotated dataset and a standard evaluation benchmark are still missing.} To deal with this issue, we set up the Automatic Detection challenge on Age-related Macular degeneration (ADAM) for the first time, held as a satellite event of the ISBI 2020 conference. The ADAM challenge consisted of four tasks which cover the main topics in detecting AMD from fundus images, including classification of AMD, detection and segmentation of optic disc, localization of fovea, and detection and segmentation of lesions. The ADAM challenge has released a comprehensive dataset of 1200 fundus images with the category labels of AMD, the pixel-wise segmentation masks of the full optic disc and lesions (drusen, exudate, hemorrhage, scar, and other), as well as the location coordinates of the macular fovea. A uniform evaluation framework has been built to make a fair comparison of different models. During the ADAM challenge, 610 results were submitted for online evaluation, and finally, 11 teams participated in the onsite challenge. This paper introduces the challenge, dataset, and evaluation methods, as well as summarizes the methods and analyzes the results of the participating teams of each task. In particular, we observed that ensembling strategy and clinical prior knowledge can better improve the performances of the deep learning models.
Learning to Detect Fake Face Images in the Wild
Oct 18, 2018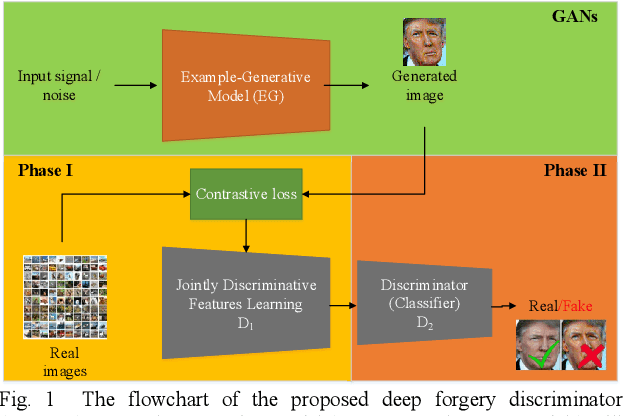
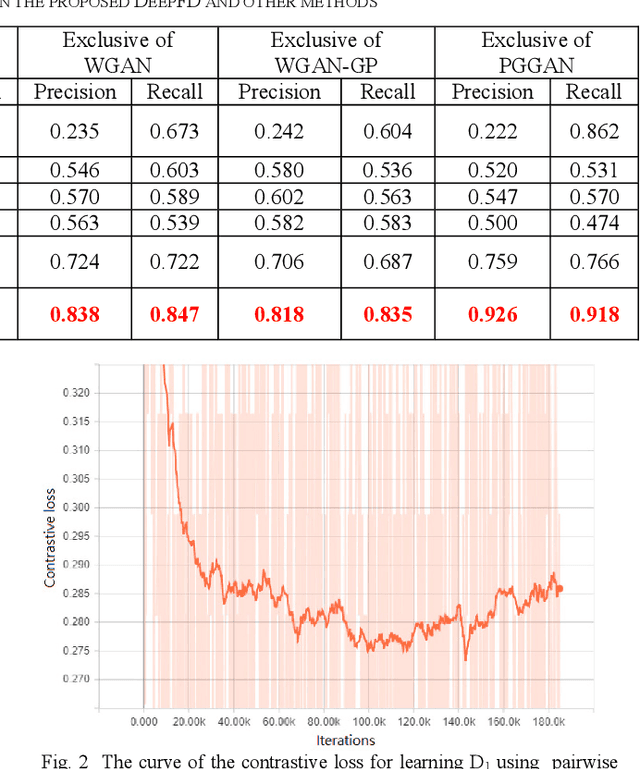
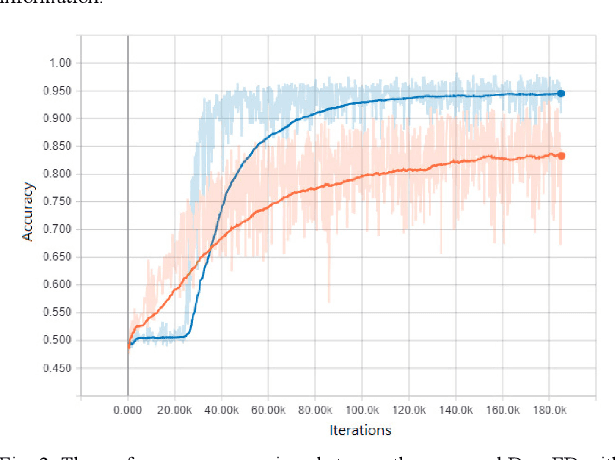

Abstract:Although Generative Adversarial Network (GAN) can be used to generate the realistic image, improper use of these technologies brings hidden concerns. For example, GAN can be used to generate a tampered video for specific people and inappropriate events, creating images that are detrimental to a particular person, and may even affect that personal safety. In this paper, we will develop a deep forgery discriminator (DeepFD) to efficiently and effectively detect the computer-generated images. Directly learning a binary classifier is relatively tricky since it is hard to find the common discriminative features for judging the fake images generated from different GANs. To address this shortcoming, we adopt contrastive loss in seeking the typical features of the synthesized images generated by different GANs and follow by concatenating a classifier to detect such computer-generated images. Experimental results demonstrate that the proposed DeepFD successfully detected 94.7% fake images generated by several state-of-the-art GANs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge