"cancer detection": models, code, and papers
Disparities in Dermatology AI: Assessments Using Diverse Clinical Images
Nov 15, 2021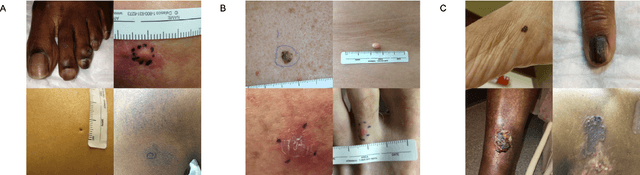

More than 3 billion people lack access to care for skin disease. AI diagnostic tools may aid in early skin cancer detection; however most models have not been assessed on images of diverse skin tones or uncommon diseases. To address this, we curated the Diverse Dermatology Images (DDI) dataset - the first publicly available, pathologically confirmed images featuring diverse skin tones. We show that state-of-the-art dermatology AI models perform substantially worse on DDI, with ROC-AUC dropping 29-40 percent compared to the models' original results. We find that dark skin tones and uncommon diseases, which are well represented in the DDI dataset, lead to performance drop-offs. Additionally, we show that state-of-the-art robust training methods cannot correct for these biases without diverse training data. Our findings identify important weaknesses and biases in dermatology AI that need to be addressed to ensure reliable application to diverse patients and across all disease.
SSD-KD: A Self-supervised Diverse Knowledge Distillation Method for Lightweight Skin Lesion Classification Using Dermoscopic Images
Mar 30, 2022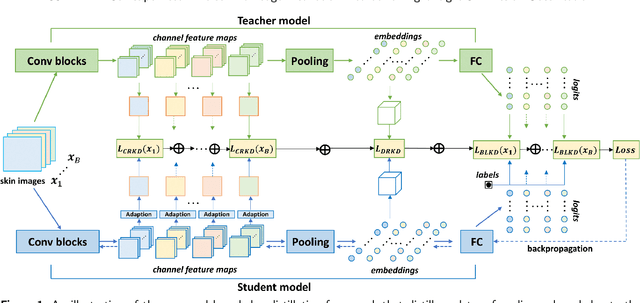

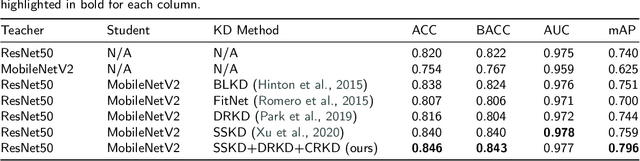
Skin cancer is one of the most common types of malignancy, affecting a large population and causing a heavy economic burden worldwide. Over the last few years, computer-aided diagnosis has been rapidly developed and make great progress in healthcare and medical practices due to the advances in artificial intelligence. However, most studies in skin cancer detection keep pursuing high prediction accuracies without considering the limitation of computing resources on portable devices. In this case, knowledge distillation (KD) has been proven as an efficient tool to help improve the adaptability of lightweight models under limited resources, meanwhile keeping a high-level representation capability. To bridge the gap, this study specifically proposes a novel method, termed SSD-KD, that unifies diverse knowledge into a generic KD framework for skin diseases classification. Our method models an intra-instance relational feature representation and integrates it with existing KD research. A dual relational knowledge distillation architecture is self-supervisedly trained while the weighted softened outputs are also exploited to enable the student model to capture richer knowledge from the teacher model. To demonstrate the effectiveness of our method, we conduct experiments on ISIC 2019, a large-scale open-accessed benchmark of skin diseases dermoscopic images. Experiments show that our distilled lightweight model can achieve an accuracy as high as 85% for the classification tasks of 8 different skin diseases with minimal parameters and computing requirements. Ablation studies confirm the effectiveness of our intra- and inter-instance relational knowledge integration strategy. Compared with state-of-the-art knowledge distillation techniques, the proposed method demonstrates improved performances for multi-diseases classification on the large-scale dermoscopy database.
Check and Link: Pairwise Lesion Correspondence Guides Mammogram Mass Detection
Sep 13, 2022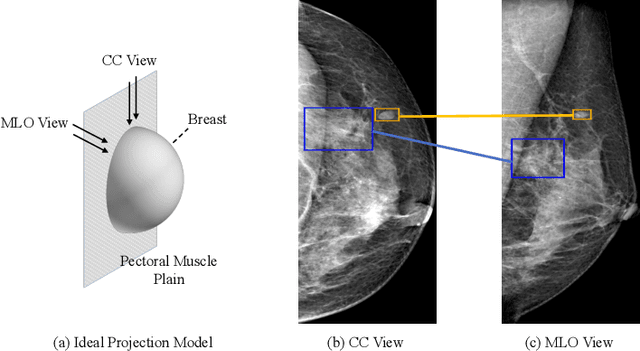
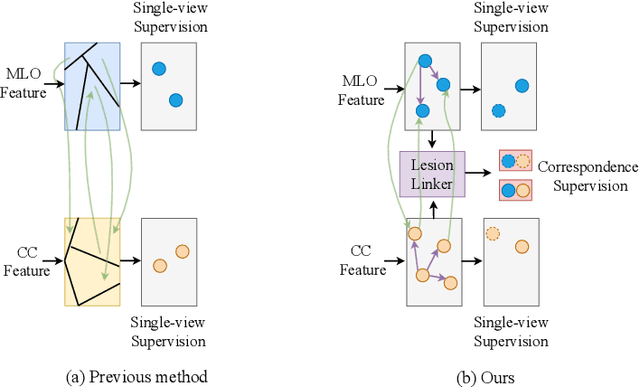
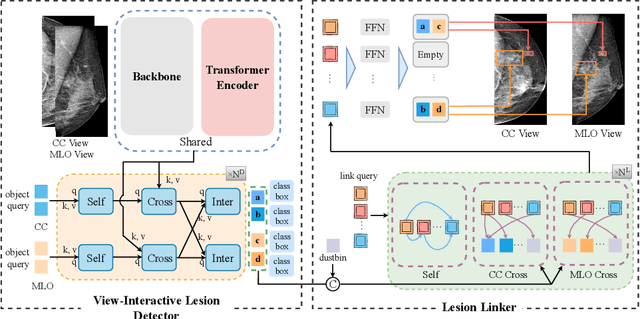
Detecting mass in mammogram is significant due to the high occurrence and mortality of breast cancer. In mammogram mass detection, modeling pairwise lesion correspondence explicitly is particularly important. However, most of the existing methods build relatively coarse correspondence and have not utilized correspondence supervision. In this paper, we propose a new transformer-based framework CL-Net to learn lesion detection and pairwise correspondence in an end-to-end manner. In CL-Net, View-Interactive Lesion Detector is proposed to achieve dynamic interaction across candidates of cross views, while Lesion Linker employs the correspondence supervision to guide the interaction process more accurately. The combination of these two designs accomplishes precise understanding of pairwise lesion correspondence for mammograms. Experiments show that CL-Net yields state-of-the-art performance on the public DDSM dataset and our in-house dataset. Moreover, it outperforms previous methods by a large margin in low FPI regime.
BI-RADS-Net: An Explainable Multitask Learning Approach for Cancer Diagnosis in Breast Ultrasound Images
Oct 05, 2021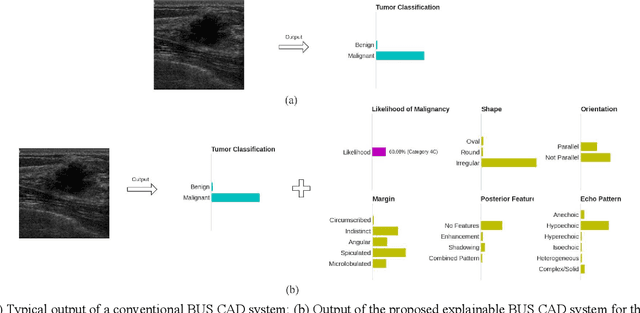
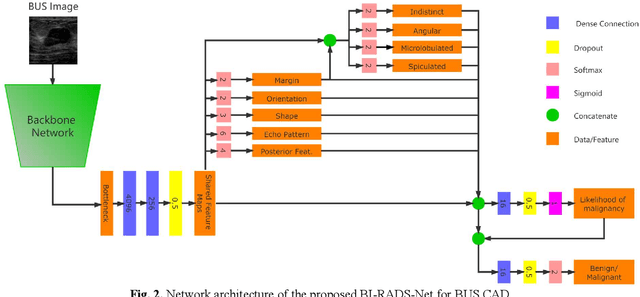
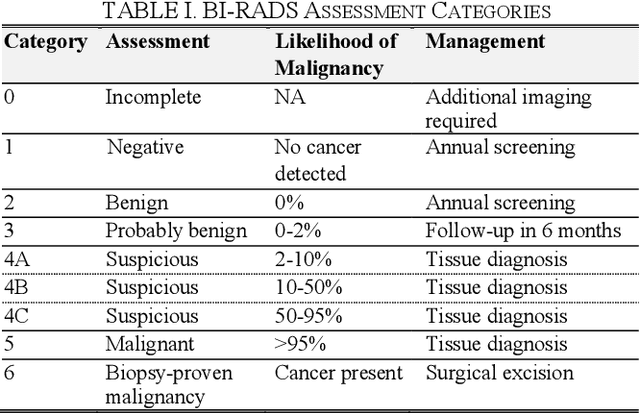
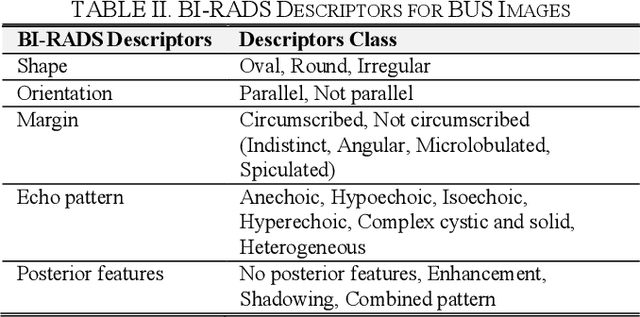
In healthcare, it is essential to explain the decision-making process of machine learning models to establish the trustworthiness of clinicians. This paper introduces BI-RADS-Net, a novel explainable deep learning approach for cancer detection in breast ultrasound images. The proposed approach incorporates tasks for explaining and classifying breast tumors, by learning feature representations relevant to clinical diagnosis. Explanations of the predictions (benign or malignant) are provided in terms of morphological features that are used by clinicians for diagnosis and reporting in medical practice. The employed features include the BI-RADS descriptors of shape, orientation, margin, echo pattern, and posterior features. Additionally, our approach predicts the likelihood of malignancy of the findings, which relates to the BI-RADS assessment category reported by clinicians. Experimental validation on a dataset consisting of 1,192 images indicates improved model accuracy, supported by explanations in clinical terms using the BI-RADS lexicon.
Properties Of Winning Tickets On Skin Lesion Classification
Aug 25, 2020
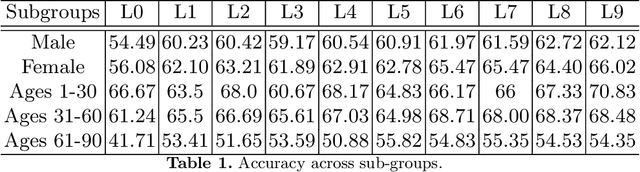
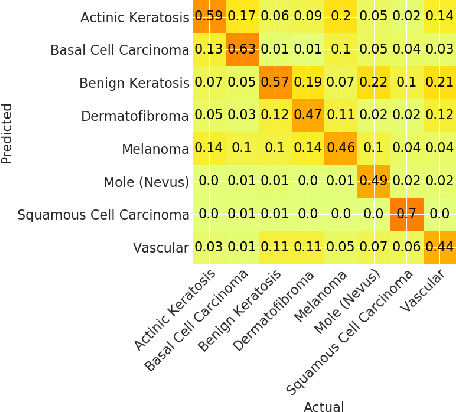
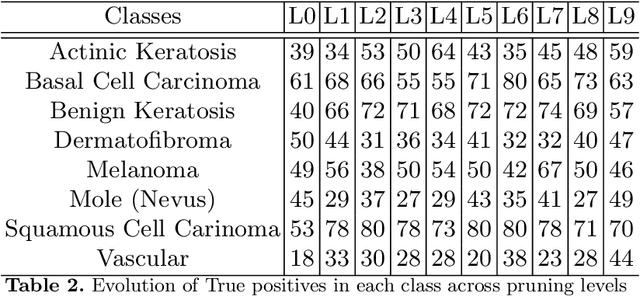
Skin cancer affects a large population every year -- automated skin cancer detection algorithms can thus greatly help clinicians. Prior efforts involving deep learning models have high detection accuracy. However, most of the models have a large number of parameters, with some works even using an ensemble of models to achieve good accuracy. In this paper, we investigate a recently proposed pruning technique called Lottery Ticket Hypothesis. We find that iterative pruning of the network resulted in improved accuracy, compared to that of the unpruned network, implying that -- the lottery ticket hypothesis can be applied to the problem of skin cancer detection and this hypothesis can result in a smaller network for inference. We also examine the accuracy across sub-groups -- created by gender and age -- and it was found that some sub-groups show a larger increase in accuracy than others.
EDEN : An Event DEtection Network for the annotation of Breast Cancer recurrences in administrative claims data
Nov 15, 2022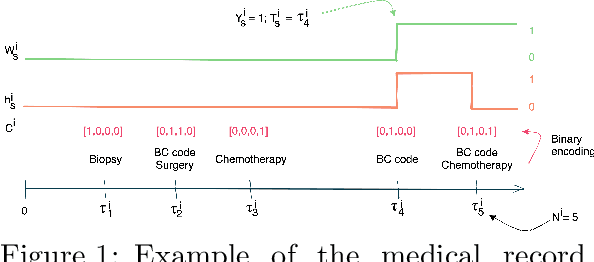
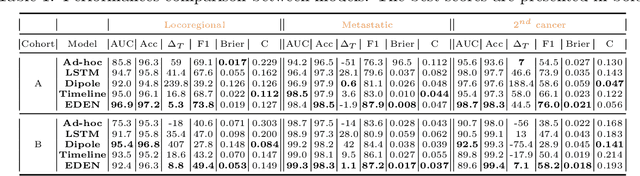
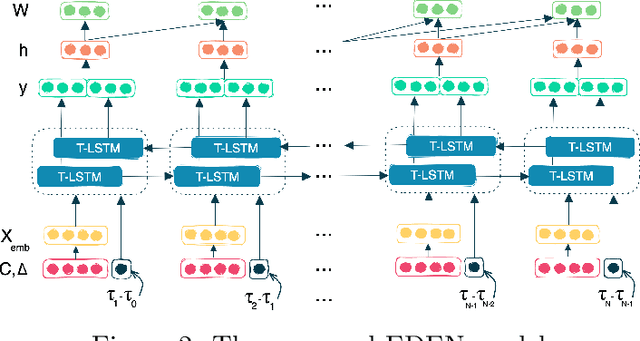
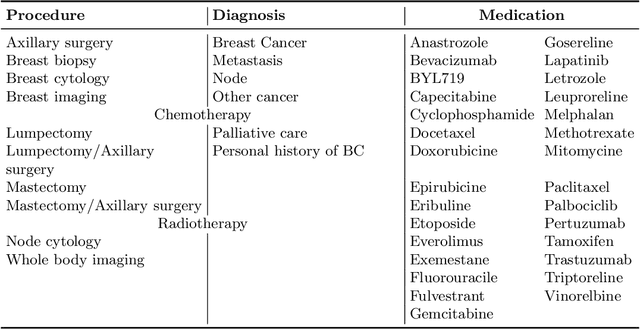
While the emergence of large administrative claims data provides opportunities for research, their use remains limited by the lack of clinical annotations relevant to disease outcomes, such as recurrence in breast cancer (BC). Several challenges arise from the annotation of such endpoints in administrative claims, including the need to infer both the occurrence and the date of the recurrence, the right-censoring of data, or the importance of time intervals between medical visits. Deep learning approaches have been successfully used to label temporal medical sequences, but no method is currently able to handle simultaneously right-censoring and visit temporality to detect survival events in medical sequences. We propose EDEN (Event DEtection Network), a time-aware Long-Short-Term-Memory network for survival analyses, and its custom loss function. Our method outperforms several state-of-the-art approaches on real-world BC datasets. EDEN constitutes a powerful tool to annotate disease recurrence from administrative claims, thus paving the way for the massive use of such data in BC research.
Ovarian Cancer Detection based on Dimensionality Reduction Techniques and Genetic Algorithm
May 04, 2021In this research, we have two serum SELDI (surface-enhanced laser desorption and ionization) mass spectra (MS) datasets to be used to select features amongst them to identify proteomic cancerous serums from normal serums. Features selection techniques have been applied and classification techniques have been applied as well. Amongst the features selection techniques we have chosen to evaluate the performance of PCA (Principal Component Analysis ) and GA (Genetic algorithm), and amongst the classification techniques we have chosen the LDA (Linear Discriminant Analysis) and Neural networks so as to evaluate the ability of the selected features in identifying the cancerous patterns. Results were obtained for two combinations of features selection techniques and classification techniques, the first one was PCA+(t-test) technique for features selection and LDA for accuracy tracking yielded an accuracy of 93.0233 % , the other one was genetic algorithm and neural network yielded an accuracy of 100%. So, we conclude that GA is more efficient for features selection and hence for cancerous patterns detection than PCA technique.
* 7 Pages
A Novel Hybrid Endoscopic Dataset for Evaluating Machine Learning-based Photometric Image Enhancement Models
Jul 06, 2022
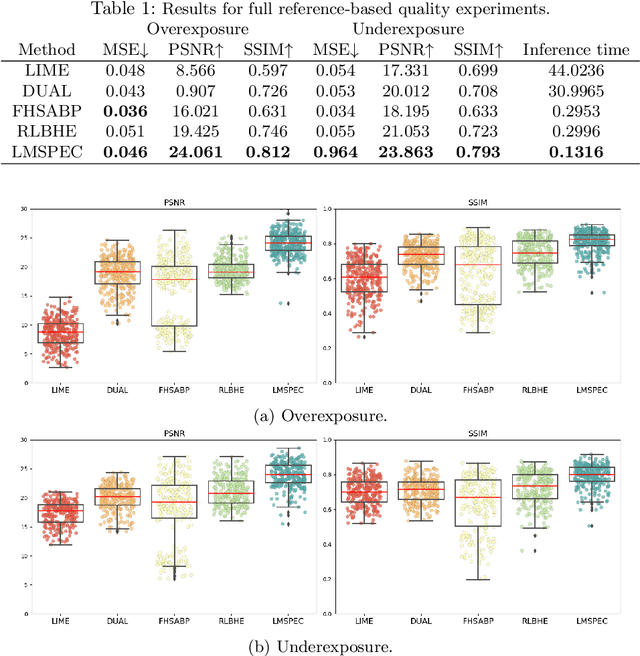
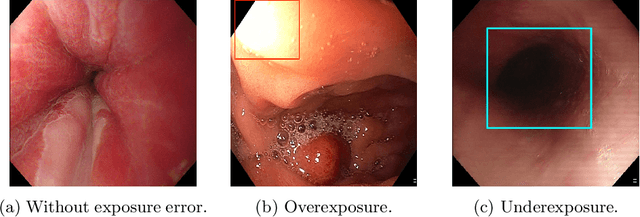
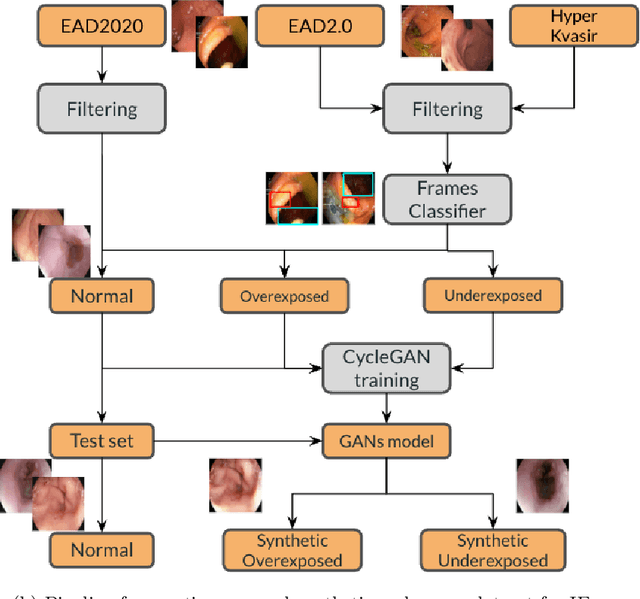
Endoscopy is the most widely used medical technique for cancer and polyp detection inside hollow organs. However, images acquired by an endoscope are frequently affected by illumination artefacts due to the enlightenment source orientation. There exist two major issues when the endoscope's light source pose suddenly changes: overexposed and underexposed tissue areas are produced. These two scenarios can result in misdiagnosis due to the lack of information in the affected zones or hamper the performance of various computer vision methods (e.g., SLAM, structure from motion, optical flow) used during the non invasive examination. The aim of this work is two-fold: i) to introduce a new synthetically generated data-set generated by a generative adversarial techniques and ii) and to explore both shallow based and deep learning-based image-enhancement methods in overexposed and underexposed lighting conditions. Best quantitative results (i.e., metric based results), were obtained by the deep-learnnig-based LMSPEC method,besides a running time around 7.6 fps)
Prostate Cancer Malignancy Detection and localization from mpMRI using auto-Deep Learning: One Step Closer to Clinical Utilization
Jun 13, 2022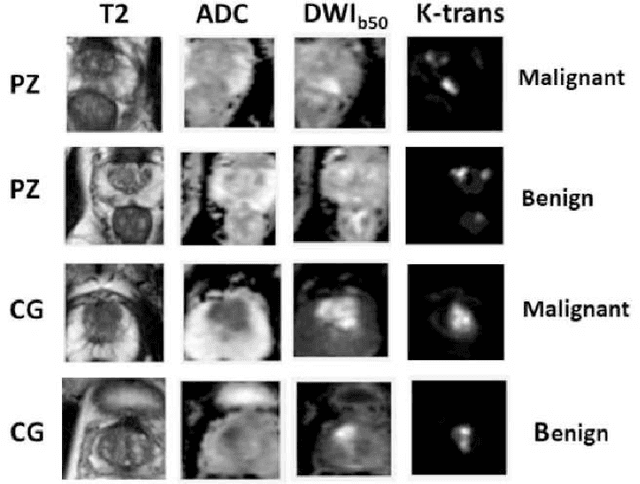

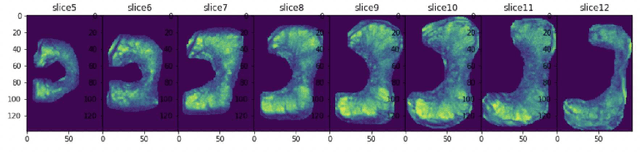
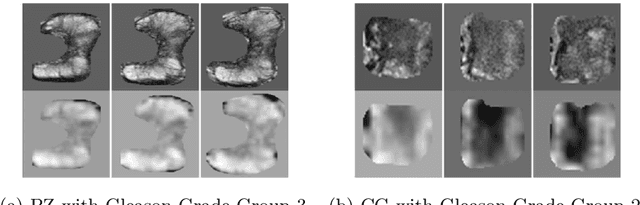
Automatic diagnosis of malignant prostate cancer patients from mpMRI has been studied heavily in the past years. Model interpretation and domain drift have been the main road blocks for clinical utilization. As an extension from our previous work where we trained a customized convolutional neural network on a public cohort with 201 patients and the cropped 2D patches around the region of interest were used as the input, the cropped 2.5D slices of the prostate glands were used as the input, and the optimal model were searched in the model space using autoKeras. Something different was peripheral zone (PZ) and central gland (CG) were trained and tested separately, the PZ detector and CG detector were demonstrated effectively in highlighting the most suspicious slices out of a sequence, hopefully to greatly ease the workload for the physicians.
Towards Reliable and Explainable AI Model for Solid Pulmonary Nodule Diagnosis
Apr 08, 2022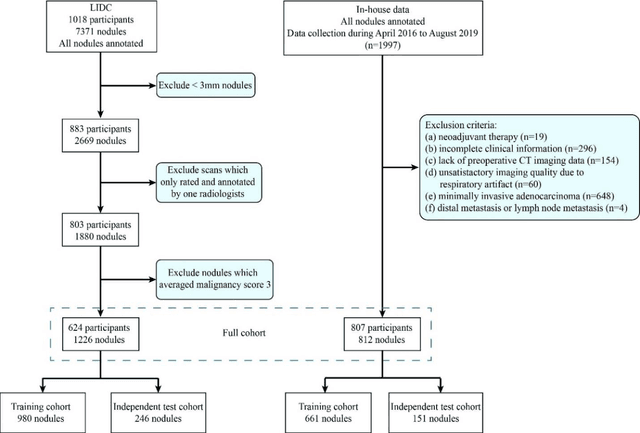
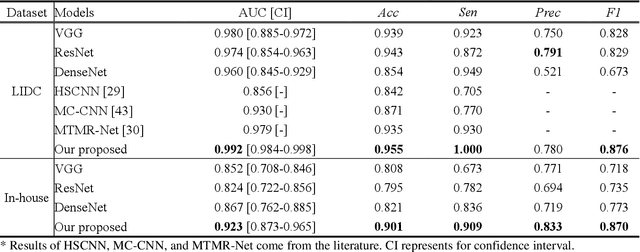
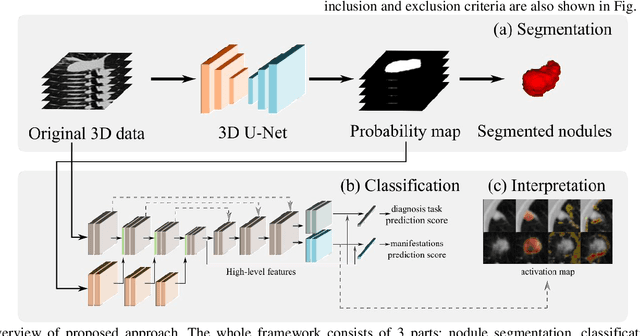
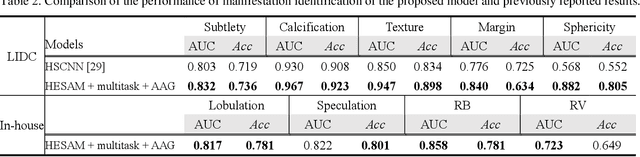
Lung cancer has the highest mortality rate of deadly cancers in the world. Early detection is essential to treatment of lung cancer. However, detection and accurate diagnosis of pulmonary nodules depend heavily on the experiences of radiologists and can be a heavy workload for them. Computer-aided diagnosis (CAD) systems have been developed to assist radiologists in nodule detection and diagnosis, greatly easing the workload while increasing diagnosis accuracy. Recent development of deep learning, greatly improved the performance of CAD systems. However, lack of model reliability and interpretability remains a major obstacle for its large-scale clinical application. In this work, we proposed a multi-task explainable deep-learning model for pulmonary nodule diagnosis. Our neural model can not only predict lesion malignancy but also identify relevant manifestations. Further, the location of each manifestation can also be visualized for visual interpretability. Our proposed neural model achieved a test AUC of 0.992 on LIDC public dataset and a test AUC of 0.923 on our in-house dataset. Moreover, our experimental results proved that by incorporating manifestation identification tasks into the multi-task model, the accuracy of the malignancy classification can also be improved. This multi-task explainable model may provide a scheme for better interaction with the radiologists in a clinical environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge