Tim K. Lee
AI-Enhanced 7-Point Checklist for Melanoma Detection Using Clinical Knowledge Graphs and Data-Driven Quantification
Jul 23, 2024



Abstract:The 7-point checklist (7PCL) is widely used in dermoscopy to identify malignant melanoma lesions needing urgent medical attention. It assigns point values to seven attributes: major attributes are worth two points each, and minor ones are worth one point each. A total score of three or higher prompts further evaluation, often including a biopsy. However, a significant limitation of current methods is the uniform weighting of attributes, which leads to imprecision and neglects their interconnections. Previous deep learning studies have treated the prediction of each attribute with the same importance as predicting melanoma, which fails to recognize the clinical significance of the attributes for melanoma. To address these limitations, we introduce a novel diagnostic method that integrates two innovative elements: a Clinical Knowledge-Based Topological Graph (CKTG) and a Gradient Diagnostic Strategy with Data-Driven Weighting Standards (GD-DDW). The CKTG integrates 7PCL attributes with diagnostic information, revealing both internal and external associations. By employing adaptive receptive domains and weighted edges, we establish connections among melanoma's relevant features. Concurrently, GD-DDW emulates dermatologists' diagnostic processes, who first observe the visual characteristics associated with melanoma and then make predictions. Our model uses two imaging modalities for the same lesion, ensuring comprehensive feature acquisition. Our method shows outstanding performance in predicting malignant melanoma and its features, achieving an average AUC value of 85%. This was validated on the EDRA dataset, the largest publicly available dataset for the 7-point checklist algorithm. Specifically, the integrated weighting system can provide clinicians with valuable data-driven benchmarks for their evaluations.
SSD-KD: A Self-supervised Diverse Knowledge Distillation Method for Lightweight Skin Lesion Classification Using Dermoscopic Images
Mar 30, 2022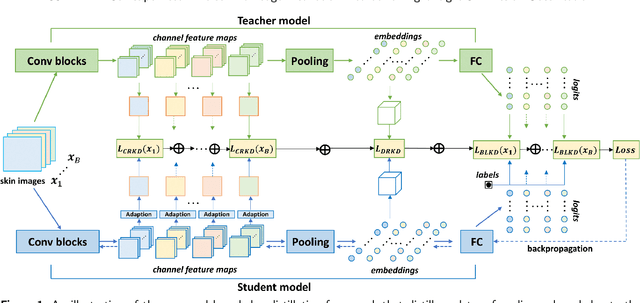

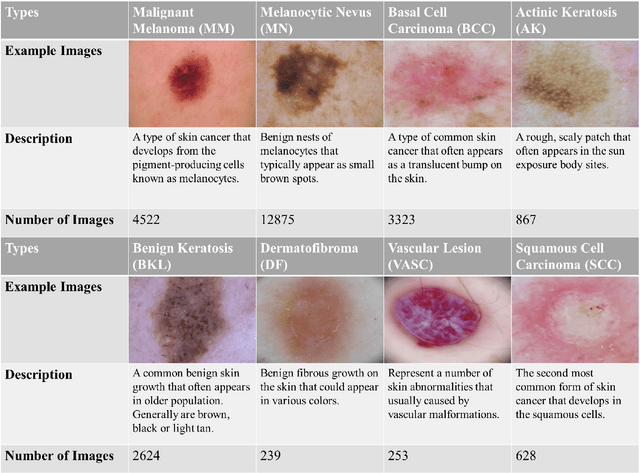
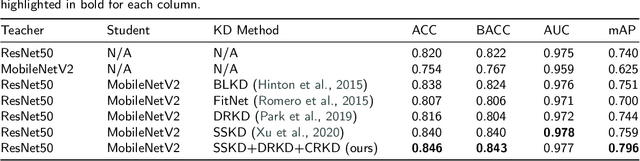
Abstract:Skin cancer is one of the most common types of malignancy, affecting a large population and causing a heavy economic burden worldwide. Over the last few years, computer-aided diagnosis has been rapidly developed and make great progress in healthcare and medical practices due to the advances in artificial intelligence. However, most studies in skin cancer detection keep pursuing high prediction accuracies without considering the limitation of computing resources on portable devices. In this case, knowledge distillation (KD) has been proven as an efficient tool to help improve the adaptability of lightweight models under limited resources, meanwhile keeping a high-level representation capability. To bridge the gap, this study specifically proposes a novel method, termed SSD-KD, that unifies diverse knowledge into a generic KD framework for skin diseases classification. Our method models an intra-instance relational feature representation and integrates it with existing KD research. A dual relational knowledge distillation architecture is self-supervisedly trained while the weighted softened outputs are also exploited to enable the student model to capture richer knowledge from the teacher model. To demonstrate the effectiveness of our method, we conduct experiments on ISIC 2019, a large-scale open-accessed benchmark of skin diseases dermoscopic images. Experiments show that our distilled lightweight model can achieve an accuracy as high as 85% for the classification tasks of 8 different skin diseases with minimal parameters and computing requirements. Ablation studies confirm the effectiveness of our intra- and inter-instance relational knowledge integration strategy. Compared with state-of-the-art knowledge distillation techniques, the proposed method demonstrates improved performances for multi-diseases classification on the large-scale dermoscopy database.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge