cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Robustness and sex differences in skin cancer detection: logistic regression vs CNNs
Apr 15, 2025



Deep learning has been reported to achieve high performances in the detection of skin cancer, yet many challenges regarding the reproducibility of results and biases remain. This study is a replication (different data, same analysis) of a study on Alzheimer's disease [28] which studied robustness of logistic regression (LR) and convolutional neural networks (CNN) across patient sexes. We explore sex bias in skin cancer detection, using the PAD-UFES-20 dataset with LR trained on handcrafted features reflecting dermatological guidelines (ABCDE and the 7-point checklist), and a pre-trained ResNet-50 model. We evaluate these models in alignment with [28]: across multiple training datasets with varied sex composition to determine their robustness. Our results show that both the LR and the CNN were robust to the sex distributions, but the results also revealed that the CNN had a significantly higher accuracy (ACC) and area under the receiver operating characteristics (AUROC) for male patients than for female patients. We hope these findings to contribute to the growing field of investigating potential bias in popular medical machine learning methods. The data and relevant scripts to reproduce our results can be found in our Github.
Towards Facilitated Fairness Assessment of AI-based Skin Lesion Classifiers Through GenAI-based Image Synthesis
Jul 23, 2025Recent advancements in Deep Learning and its application on the edge hold great potential for the revolution of routine screenings for skin cancers like Melanoma. Along with the anticipated benefits of this technology, potential dangers arise from unforseen and inherent biases. Thus, assessing and improving the fairness of such systems is of utmost importance. A key challenge in fairness assessment is to ensure that the evaluation dataset is sufficiently representative of different Personal Identifiable Information (PII) (sex, age, and race) and other minority groups. Against the backdrop of this challenge, this study leverages the state-of-the-art Generative AI (GenAI) LightningDiT model to assess the fairness of publicly available melanoma classifiers. The results suggest that fairness assessment using highly realistic synthetic data is a promising direction. Yet, our findings indicate that verifying fairness becomes difficult when the melanoma-detection model used for evaluation is trained on data that differ from the dataset underpinning the synthetic images. Nonetheless, we propose that our approach offers a valuable new avenue for employing synthetic data to gauge and enhance fairness in medical-imaging GenAI systems.
Vision-Language Model-Based Semantic-Guided Imaging Biomarker for Early Lung Cancer Detection
Apr 30, 2025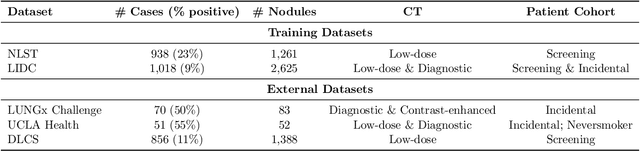
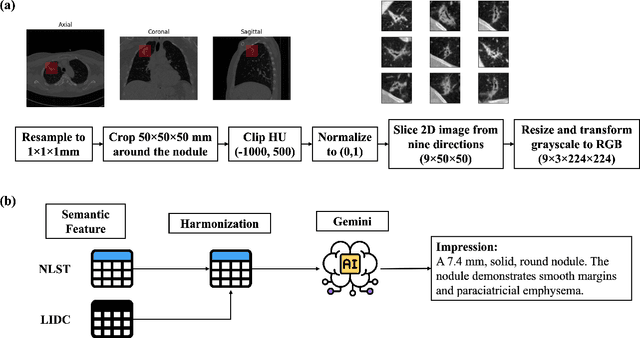
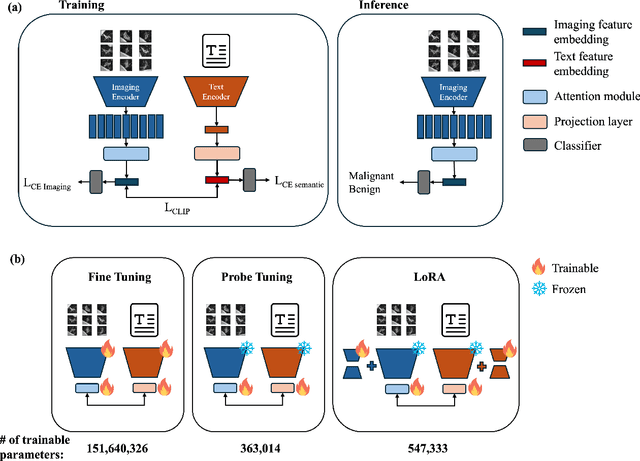
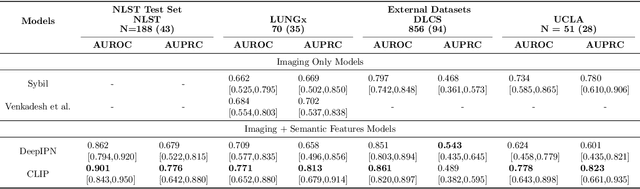
Objective: A number of machine learning models have utilized semantic features, deep features, or both to assess lung nodule malignancy. However, their reliance on manual annotation during inference, limited interpretability, and sensitivity to imaging variations hinder their application in real-world clinical settings. Thus, this research aims to integrate semantic features derived from radiologists' assessments of nodules, allowing the model to learn clinically relevant, robust, and explainable features for predicting lung cancer. Methods: We obtained 938 low-dose CT scans from the National Lung Screening Trial with 1,246 nodules and semantic features. The Lung Image Database Consortium dataset contains 1,018 CT scans, with 2,625 lesions annotated for nodule characteristics. Three external datasets were obtained from UCLA Health, the LUNGx Challenge, and the Duke Lung Cancer Screening. We finetuned a pretrained Contrastive Language-Image Pretraining model with a parameter-efficient fine-tuning approach to align imaging and semantic features and predict the one-year lung cancer diagnosis. Results: We evaluated the performance of the one-year diagnosis of lung cancer with AUROC and AUPRC and compared it to three state-of-the-art models. Our model demonstrated an AUROC of 0.90 and AUPRC of 0.78, outperforming baseline state-of-the-art models on external datasets. Using CLIP, we also obtained predictions on semantic features, such as nodule margin (AUROC: 0.81), nodule consistency (0.81), and pleural attachment (0.84), that can be used to explain model predictions. Conclusion: Our approach accurately classifies lung nodules as benign or malignant, providing explainable outputs, aiding clinicians in comprehending the underlying meaning of model predictions. This approach also prevents the model from learning shortcuts and generalizes across clinical settings.
BreastDCEDL: Curating a Comprehensive DCE-MRI Dataset and developing a Transformer Implementation for Breast Cancer Treatment Response Prediction
Jun 13, 2025Breast cancer remains a leading cause of cancer-related mortality worldwide, making early detection and accurate treatment response monitoring critical priorities. We present BreastDCEDL, a curated, deep learning-ready dataset comprising pre-treatment 3D Dynamic Contrast-Enhanced MRI (DCE-MRI) scans from 2,070 breast cancer patients drawn from the I-SPY1, I-SPY2, and Duke cohorts, all sourced from The Cancer Imaging Archive. The raw DICOM imaging data were rigorously converted into standardized 3D NIfTI volumes with preserved signal integrity, accompanied by unified tumor annotations and harmonized clinical metadata including pathologic complete response (pCR), hormone receptor (HR), and HER2 status. Although DCE-MRI provides essential diagnostic information and deep learning offers tremendous potential for analyzing such complex data, progress has been limited by lack of accessible, public, multicenter datasets. BreastDCEDL addresses this gap by enabling development of advanced models, including state-of-the-art transformer architectures that require substantial training data. To demonstrate its capacity for robust modeling, we developed the first transformer-based model for breast DCE-MRI, leveraging Vision Transformer (ViT) architecture trained on RGB-fused images from three contrast phases (pre-contrast, early post-contrast, and late post-contrast). Our ViT model achieved state-of-the-art pCR prediction performance in HR+/HER2- patients (AUC 0.94, accuracy 0.93). BreastDCEDL includes predefined benchmark splits, offering a framework for reproducible research and enabling clinically meaningful modeling in breast cancer imaging.
White Light Specular Reflection Data Augmentation for Deep Learning Polyp Detection
May 08, 2025Colorectal cancer is one of the deadliest cancers today, but it can be prevented through early detection of malignant polyps in the colon, primarily via colonoscopies. While this method has saved many lives, human error remains a significant challenge, as missing a polyp could have fatal consequences for the patient. Deep learning (DL) polyp detectors offer a promising solution. However, existing DL polyp detectors often mistake white light reflections from the endoscope for polyps, which can lead to false positives.To address this challenge, in this paper, we propose a novel data augmentation approach that artificially adds more white light reflections to create harder training scenarios. Specifically, we first generate a bank of artificial lights using the training dataset. Then we find the regions of the training images that we should not add these artificial lights on. Finally, we propose a sliding window method to add the artificial light to the areas that fit of the training images, resulting in augmented images. By providing the model with more opportunities to make mistakes, we hypothesize that it will also have more chances to learn from those mistakes, ultimately improving its performance in polyp detection. Experimental results demonstrate the effectiveness of our new data augmentation method.
A Comprehensive Study on Medical Image Segmentation using Deep Neural Networks
Jun 04, 2025Over the past decade, Medical Image Segmentation (MIS) using Deep Neural Networks (DNNs) has achieved significant performance improvements and holds great promise for future developments. This paper presents a comprehensive study on MIS based on DNNs. Intelligent Vision Systems are often evaluated based on their output levels, such as Data, Information, Knowledge, Intelligence, and Wisdom (DIKIW),and the state-of-the-art solutions in MIS at these levels are the focus of research. Additionally, Explainable Artificial Intelligence (XAI) has become an important research direction, as it aims to uncover the "black box" nature of previous DNN architectures to meet the requirements of transparency and ethics. The study emphasizes the importance of MIS in disease diagnosis and early detection, particularly for increasing the survival rate of cancer patients through timely diagnosis. XAI and early prediction are considered two important steps in the journey from "intelligence" to "wisdom." Additionally, the paper addresses existing challenges and proposes potential solutions to enhance the efficiency of implementing DNN-based MIS.
Model-Independent Machine Learning Approach for Nanometric Axial Localization and Tracking
May 20, 2025Accurately tracking particles and determining their position along the optical axis is a major challenge in optical microscopy, especially when extremely high precision is needed. In this study, we introduce a deep learning approach using convolutional neural networks (CNNs) that can determine axial positions from dual-focal plane images without relying on predefined models. Our method achieves an axial localization accuracy of 40 nanometers - six times better than traditional single-focal plane techniques. The model's simple design and strong performance make it suitable for a wide range of uses, including dark matter detection, proton therapy for cancer, and radiation protection in space. It also shows promise in fields like biological imaging, materials science, and environmental monitoring. This work highlights how machine learning can turn complex image data into reliable, precise information, offering a flexible and powerful tool for many scientific applications.
Discovering Pathology Rationale and Token Allocation for Efficient Multimodal Pathology Reasoning
May 21, 2025Multimodal pathological image understanding has garnered widespread interest due to its potential to improve diagnostic accuracy and enable personalized treatment through integrated visual and textual data. However, existing methods exhibit limited reasoning capabilities, which hamper their ability to handle complex diagnostic scenarios. Additionally, the enormous size of pathological images leads to severe computational burdens, further restricting their practical deployment. To address these limitations, we introduce a novel bilateral reinforcement learning framework comprising two synergistic branches. One reinforcement branch enhances the reasoning capability by enabling the model to learn task-specific decision processes, i.e., pathology rationales, directly from labels without explicit reasoning supervision. While the other branch dynamically allocates a tailored number of tokens to different images based on both their visual content and task context, thereby optimizing computational efficiency. We apply our method to various pathological tasks such as visual question answering, cancer subtyping, and lesion detection. Extensive experiments show an average +41.7 absolute performance improvement with 70.3% lower inference costs over the base models, achieving both reasoning accuracy and computational efficiency.
Lung Nodule-SSM: Self-Supervised Lung Nodule Detection and Classification in Thoracic CT Images
May 21, 2025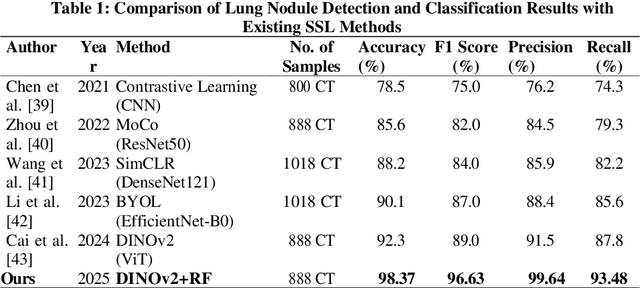
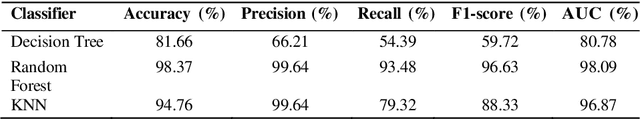
Lung cancer remains among the deadliest types of cancer in recent decades, and early lung nodule detection is crucial for improving patient outcomes. The limited availability of annotated medical imaging data remains a bottleneck in developing accurate computer-aided diagnosis (CAD) systems. Self-supervised learning can help leverage large amounts of unlabeled data to develop more robust CAD systems. With the recent advent of transformer-based architecture and their ability to generalize to unseen tasks, there has been an effort within the healthcare community to adapt them to various medical downstream tasks. Thus, we propose a novel "LungNodule-SSM" method, which utilizes selfsupervised learning with DINOv2 as a backbone to enhance lung nodule detection and classification without annotated data. Our methodology has two stages: firstly, the DINOv2 model is pre-trained on unlabeled CT scans to learn robust feature representations, then secondly, these features are fine-tuned using transformer-based architectures for lesionlevel detection and accurate lung nodule diagnosis. The proposed method has been evaluated on the challenging LUNA 16 dataset, consisting of 888 CT scans, and compared with SOTA methods. Our experimental results show the superiority of our proposed method with an accuracy of 98.37%, explaining its effectiveness in lung nodule detection. The source code, datasets, and pre-processed data can be accessed using the link:https://github.com/EMeRALDsNRPU/Lung-Nodule-SSM-Self-Supervised-Lung-Nodule-Detection-and-Classification/tree/main
Automated Detection of Clinical Entities in Lung and Breast Cancer Reports Using NLP Techniques
May 14, 2025Research projects, including those focused on cancer, rely on the manual extraction of information from clinical reports. This process is time-consuming and prone to errors, limiting the efficiency of data-driven approaches in healthcare. To address these challenges, Natural Language Processing (NLP) offers an alternative for automating the extraction of relevant data from electronic health records (EHRs). In this study, we focus on lung and breast cancer due to their high incidence and the significant impact they have on public health. Early detection and effective data management in both types of cancer are crucial for improving patient outcomes. To enhance the accuracy and efficiency of data extraction, we utilized GMV's NLP tool uQuery, which excels at identifying relevant entities in clinical texts and converting them into standardized formats such as SNOMED and OMOP. uQuery not only detects and classifies entities but also associates them with contextual information, including negated entities, temporal aspects, and patient-related details. In this work, we explore the use of NLP techniques, specifically Named Entity Recognition (NER), to automatically identify and extract key clinical information from EHRs related to these two cancers. A dataset from Health Research Institute Hospital La Fe (IIS La Fe), comprising 200 annotated breast cancer and 400 lung cancer reports, was used, with eight clinical entities manually labeled using the Doccano platform. To perform NER, we fine-tuned the bsc-bio-ehr-en3 model, a RoBERTa-based biomedical linguistic model pre-trained in Spanish. Fine-tuning was performed using the Transformers architecture, enabling accurate recognition of clinical entities in these cancer types. Our results demonstrate strong overall performance, particularly in identifying entities like MET and PAT, although challenges remain with less frequent entities like EVOL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge