Multi Modal Document Classification
Papers and Code
Towards Scalable and Cross-Lingual Specialist Language Models for Oncology
Mar 11, 2025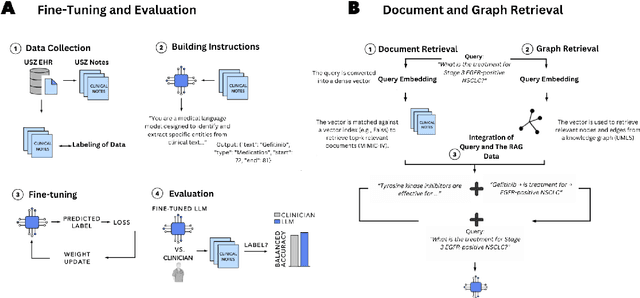
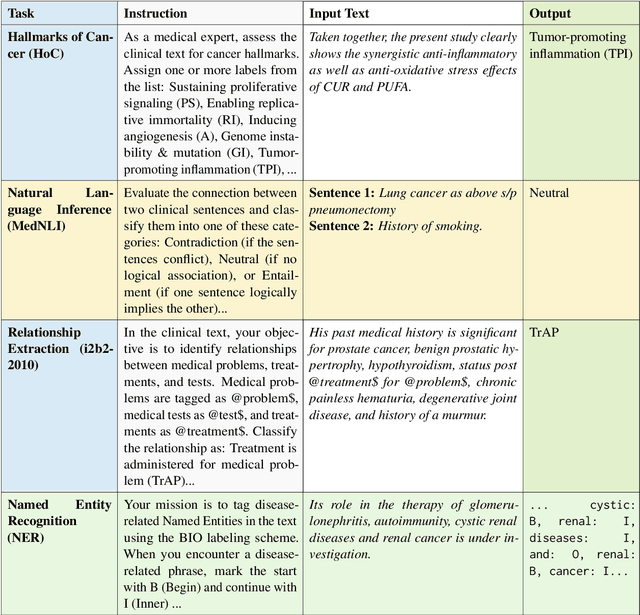
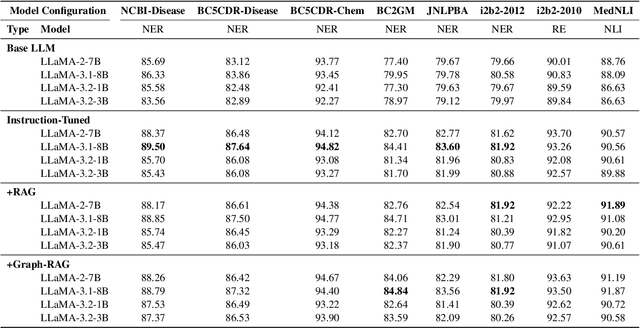
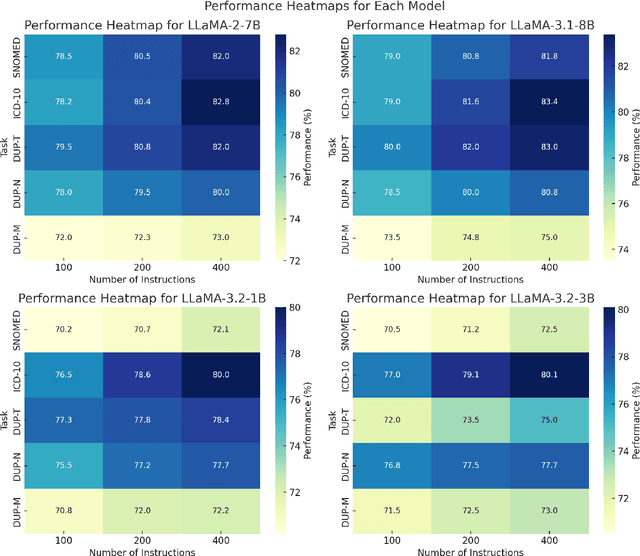
Clinical oncology generates vast, unstructured data that often contain inconsistencies, missing information, and ambiguities, making it difficult to extract reliable insights for data-driven decision-making. General-purpose large language models (LLMs) struggle with these challenges due to their lack of domain-specific reasoning, including specialized clinical terminology, context-dependent interpretations, and multi-modal data integration. We address these issues with an oncology-specialized, efficient, and adaptable NLP framework that combines instruction tuning, retrieval-augmented generation (RAG), and graph-based knowledge integration. Our lightweight models prove effective at oncology-specific tasks, such as named entity recognition (e.g., identifying cancer diagnoses), entity linking (e.g., linking entities to standardized ontologies), TNM staging, document classification (e.g., cancer subtype classification from pathology reports), and treatment response prediction. Our framework emphasizes adaptability and resource efficiency. We include minimal German instructions, collected at the University Hospital Zurich (USZ), to test whether small amounts of non-English language data can effectively transfer knowledge across languages. This approach mirrors our motivation for lightweight models, which balance strong performance with reduced computational costs, making them suitable for resource-limited healthcare settings. We validated our models on oncology datasets, demonstrating strong results in named entity recognition, relation extraction, and document classification.
Predicting Cardiopulmonary Exercise Testing Outcomes in Congenital Heart Disease Through Multi-modal Data Integration and Geometric Learning
Mar 18, 2025



Cardiopulmonary exercise testing (CPET) provides a comprehensive assessment of functional capacity by measuring key physiological variables including oxygen consumption ($VO_2$), carbon dioxide production ($VCO_2$), and pulmonary ventilation ($VE$) during exercise. Previous research has established that parameters such as peak $VO_2$ and $VE/VCO_2$ ratio serve as robust predictors of mortality risk in chronic heart failure patients. In this study, we leverage CPET variables as surrogate mortality endpoints for patients with Congenital Heart Disease (CHD). To our knowledge, this represents the first successful implementation of an advanced machine learning approach that predicts CPET outcomes by integrating electrocardiograms (ECGs) with information derived from clinical letters. Our methodology began with extracting unstructured patient information-including intervention history, diagnoses, and medication regimens-from clinical letters using natural language processing techniques, organizing this data into a structured database. We then digitized ECGs to obtain quantifiable waveforms and established comprehensive data linkages. The core innovation of our approach lies in exploiting the Riemannian geometric properties of covariance matrices derived from both 12-lead ECGs and clinical text data to develop robust regression and classification models. Through extensive ablation studies, we demonstrated that the integration of ECG signals with clinical documentation, enhanced by covariance augmentation techniques in Riemannian space, consistently produced superior predictive performance compared to conventional approaches.
WordVIS: A Color Worth A Thousand Words
Dec 13, 2024



Document classification is considered a critical element in automated document processing systems. In recent years multi-modal approaches have become increasingly popular for document classification. Despite their improvements, these approaches are underutilized in the industry due to their requirement for a tremendous volume of training data and extensive computational power. In this paper, we attempt to address these issues by embedding textual features directly into the visual space, allowing lightweight image-based classifiers to achieve state-of-the-art results using small-scale datasets in document classification. To evaluate the efficacy of the visual features generated from our approach on limited data, we tested on the standard dataset Tobacco-3482. Our experiments show a tremendous improvement in image-based classifiers, achieving an improvement of 4.64% using ResNet50 with no document pre-training. It also sets a new record for the best accuracy of the Tobacco-3482 dataset with a score of 91.14% using the image-based DocXClassifier with no document pre-training. The simplicity of the approach, its resource requirements, and subsequent results provide a good prospect for its use in industrial use cases.
Hierarchical Multi-modal Transformer for Cross-modal Long Document Classification
Jul 14, 2024



Long Document Classification (LDC) has gained significant attention recently. However, multi-modal data in long documents such as texts and images are not being effectively utilized. Prior studies in this area have attempted to integrate texts and images in document-related tasks, but they have only focused on short text sequences and images of pages. How to classify long documents with hierarchical structure texts and embedding images is a new problem and faces multi-modal representation difficulties. In this paper, we propose a novel approach called Hierarchical Multi-modal Transformer (HMT) for cross-modal long document classification. The HMT conducts multi-modal feature interaction and fusion between images and texts in a hierarchical manner. Our approach uses a multi-modal transformer and a dynamic multi-scale multi-modal transformer to model the complex relationships between image features, and the section and sentence features. Furthermore, we introduce a new interaction strategy called the dynamic mask transfer module to integrate these two transformers by propagating features between them. To validate our approach, we conduct cross-modal LDC experiments on two newly created and two publicly available multi-modal long document datasets, and the results show that the proposed HMT outperforms state-of-the-art single-modality and multi-modality methods.
Out-of-Distribution Detection with Attention Head Masking for Multimodal Document Classification
Aug 20, 2024



Detecting out-of-distribution (OOD) data is crucial in machine learning applications to mitigate the risk of model overconfidence, thereby enhancing the reliability and safety of deployed systems. The majority of existing OOD detection methods predominantly address uni-modal inputs, such as images or texts. In the context of multi-modal documents, there is a notable lack of extensive research on the performance of these methods, which have primarily been developed with a focus on computer vision tasks. We propose a novel methodology termed as attention head masking (AHM) for multi-modal OOD tasks in document classification systems. Our empirical results demonstrate that the proposed AHM method outperforms all state-of-the-art approaches and significantly decreases the false positive rate (FPR) compared to existing solutions up to 7.5\%. This methodology generalizes well to multi-modal data, such as documents, where visual and textual information are modeled under the same Transformer architecture. To address the scarcity of high-quality publicly available document datasets and encourage further research on OOD detection for documents, we introduce FinanceDocs, a new document AI dataset. Our code and dataset are publicly available.
Deep BI-RADS Network for Improved Cancer Detection from Mammograms
Nov 16, 2024While state-of-the-art models for breast cancer detection leverage multi-view mammograms for enhanced diagnostic accuracy, they often focus solely on visual mammography data. However, radiologists document valuable lesion descriptors that contain additional information that can enhance mammography-based breast cancer screening. A key question is whether deep learning models can benefit from these expert-derived features. To address this question, we introduce a novel multi-modal approach that combines textual BI-RADS lesion descriptors with visual mammogram content. Our method employs iterative attention layers to effectively fuse these different modalities, significantly improving classification performance over image-only models. Experiments on the CBIS-DDSM dataset demonstrate substantial improvements across all metrics, demonstrating the contribution of handcrafted features to end-to-end.
FinEmbedDiff: A Cost-Effective Approach of Classifying Financial Documents with Vector Sampling using Multi-modal Embedding Models
May 28, 2024



Accurate classification of multi-modal financial documents, containing text, tables, charts, and images, is crucial but challenging. Traditional text-based approaches often fail to capture the complex multi-modal nature of these documents. We propose FinEmbedDiff, a cost-effective vector sampling method that leverages pre-trained multi-modal embedding models to classify financial documents. Our approach generates multi-modal embedding vectors for documents, and compares new documents with pre-computed class embeddings using vector similarity measures. Evaluated on a large dataset, FinEmbedDiff achieves competitive classification accuracy compared to state-of-the-art baselines while significantly reducing computational costs. The method exhibits strong generalization capabilities, making it a practical and scalable solution for real-world financial applications.
* 10 pages, 3 figures
FungiTastic: A multi-modal dataset and benchmark for image categorization
Aug 24, 2024



We introduce a new, highly challenging benchmark and a dataset -- FungiTastic -- based on data continuously collected over a twenty-year span. The dataset originates in fungal records labeled and curated by experts. It consists of about 350k multi-modal observations that include more than 650k photographs from 5k fine-grained categories and diverse accompanying information, e.g., acquisition metadata, satellite images, and body part segmentation. FungiTastic is the only benchmark that includes a test set with partially DNA-sequenced ground truth of unprecedented label reliability. The benchmark is designed to support (i) standard close-set classification, (ii) open-set classification, (iii) multi-modal classification, (iv) few-shot learning, (v) domain shift, and many more. We provide baseline methods tailored for almost all the use-cases. We provide a multitude of ready-to-use pre-trained models on HuggingFace and a framework for model training. A comprehensive documentation describing the dataset features and the baselines are available at https://bohemianvra.github.io/FungiTastic/ and https://www.kaggle.com/datasets/picekl/fungitastic.
DocGenome: An Open Large-scale Scientific Document Benchmark for Training and Testing Multi-modal Large Language Models
Jun 17, 2024



Scientific documents record research findings and valuable human knowledge, comprising a vast corpus of high-quality data. Leveraging multi-modality data extracted from these documents and assessing large models' abilities to handle scientific document-oriented tasks is therefore meaningful. Despite promising advancements, large models still perform poorly on multi-page scientific document extraction and understanding tasks, and their capacity to process within-document data formats such as charts and equations remains under-explored. To address these issues, we present DocGenome, a structured document benchmark constructed by annotating 500K scientific documents from 153 disciplines in the arXiv open-access community, using our custom auto-labeling pipeline. DocGenome features four key characteristics: 1) Completeness: It is the first dataset to structure data from all modalities including 13 layout attributes along with their LaTeX source codes. 2) Logicality: It provides 6 logical relationships between different entities within each scientific document. 3) Diversity: It covers various document-oriented tasks, including document classification, visual grounding, document layout detection, document transformation, open-ended single-page QA and multi-page QA. 4) Correctness: It undergoes rigorous quality control checks conducted by a specialized team. We conduct extensive experiments to demonstrate the advantages of DocGenome and objectively evaluate the performance of large models on our benchmark.
BuDDIE: A Business Document Dataset for Multi-task Information Extraction
Apr 05, 2024The field of visually rich document understanding (VRDU) aims to solve a multitude of well-researched NLP tasks in a multi-modal domain. Several datasets exist for research on specific tasks of VRDU such as document classification (DC), key entity extraction (KEE), entity linking, visual question answering (VQA), inter alia. These datasets cover documents like invoices and receipts with sparse annotations such that they support one or two co-related tasks (e.g., entity extraction and entity linking). Unfortunately, only focusing on a single specific of documents or task is not representative of how documents often need to be processed in the wild - where variety in style and requirements is expected. In this paper, we introduce BuDDIE (Business Document Dataset for Information Extraction), the first multi-task dataset of 1,665 real-world business documents that contains rich and dense annotations for DC, KEE, and VQA. Our dataset consists of publicly available business entity documents from US state government websites. The documents are structured and vary in their style and layout across states and types (e.g., forms, certificates, reports, etc.). We provide data variety and quality metrics for BuDDIE as well as a series of baselines for each task. Our baselines cover traditional textual, multi-modal, and large language model approaches to VRDU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge