Colorectal Gland Segmentation
Papers and Code
Gland segmentation via dual encoders and boundary-enhanced attention
Jan 29, 2024



Accurate and automated gland segmentation on pathological images can assist pathologists in diagnosing the malignancy of colorectal adenocarcinoma. However, due to various gland shapes, severe deformation of malignant glands, and overlapping adhesions between glands. Gland segmentation has always been very challenging. To address these problems, we propose a DEA model. This model consists of two branches: the backbone encoding and decoding network and the local semantic extraction network. The backbone encoding and decoding network extracts advanced Semantic features, uses the proposed feature decoder to restore feature space information, and then enhances the boundary features of the gland through boundary enhancement attention. The local semantic extraction network uses the pre-trained DeepLabv3+ as a Local semantic-guided encoder to realize the extraction of edge features. Experimental results on two public datasets, GlaS and CRAG, confirm that the performance of our method is better than other gland segmentation methods.
Synthesis of Annotated Colorectal Cancer Tissue Images from Gland Layout
May 08, 2023Generating annotated pairs of realistic tissue images along with their annotations is a challenging task in computational histopathology. Such synthetic images and their annotations can be useful in training and evaluation of algorithms in the domain of computational pathology. To address this, we present an interactive framework to generate pairs of realistic colorectal cancer histology images with corresponding tissue component masks from the input gland layout. The framework shows the ability to generate realistic qualitative tissue images preserving morphological characteristics including stroma, goblet cells and glandular lumen. We show the appearance of glands can be controlled by user inputs such as number of glands, their locations and sizes. We also validate the quality of generated annotated pair with help of the gland segmentation algorithm.
One Model is All You Need: Multi-Task Learning Enables Simultaneous Histology Image Segmentation and Classification
Feb 28, 2022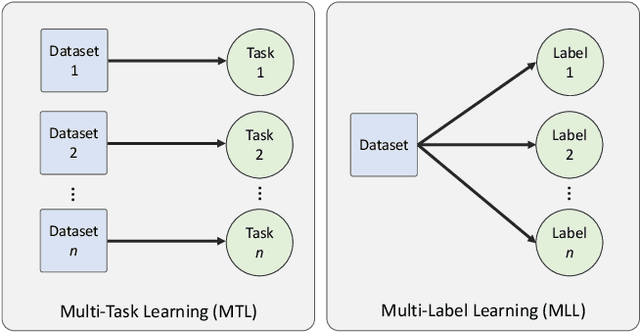
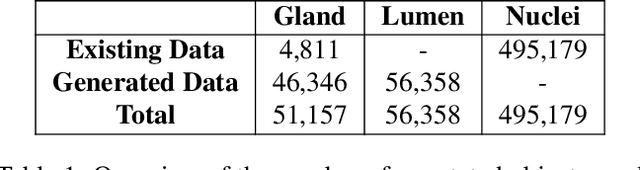
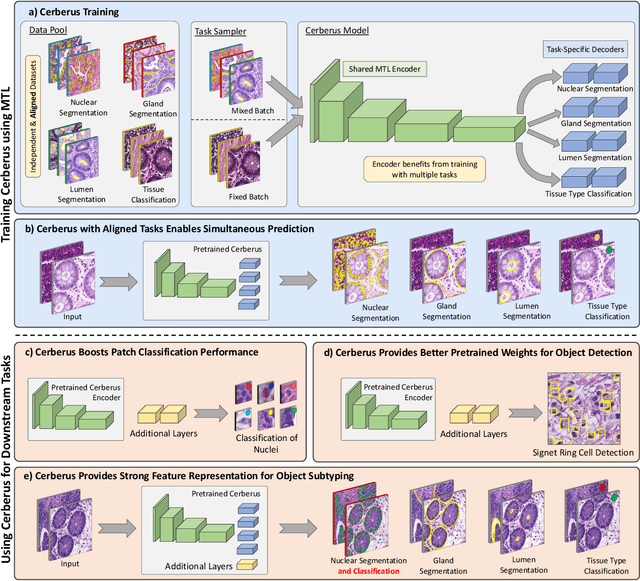
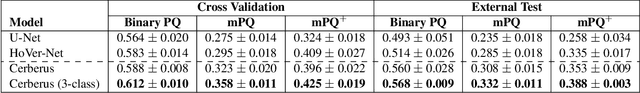
The recent surge in performance for image analysis of digitised pathology slides can largely be attributed to the advance of deep learning. Deep models can be used to initially localise various structures in the tissue and hence facilitate the extraction of interpretable features for biomarker discovery. However, these models are typically trained for a single task and therefore scale poorly as we wish to adapt the model for an increasing number of different tasks. Also, supervised deep learning models are very data hungry and therefore rely on large amounts of training data to perform well. In this paper we present a multi-task learning approach for segmentation and classification of nuclei, glands, lumen and different tissue regions that leverages data from multiple independent data sources. While ensuring that our tasks are aligned by the same tissue type and resolution, we enable simultaneous prediction with a single network. As a result of feature sharing, we also show that the learned representation can be used to improve downstream tasks, including nuclear classification and signet ring cell detection. As part of this work, we use a large dataset consisting of over 600K objects for segmentation and 440K patches for classification and make the data publicly available. We use our approach to process the colorectal subset of TCGA, consisting of 599 whole-slide images, to localise 377 million, 900K and 2.1 million nuclei, glands and lumen respectively. We make this resource available to remove a major barrier in the development of explainable models for computational pathology.
SAFRON: Stitching Across the Frontier for Generating Colorectal Cancer Histology Images
Aug 11, 2020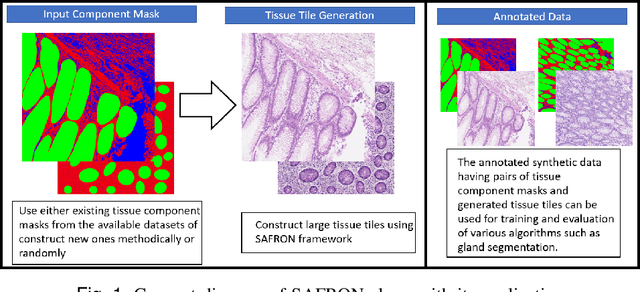
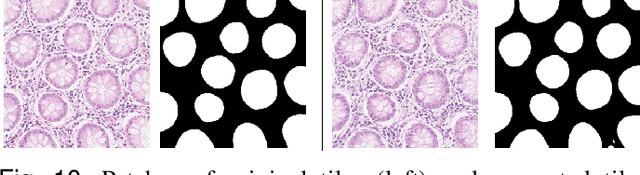
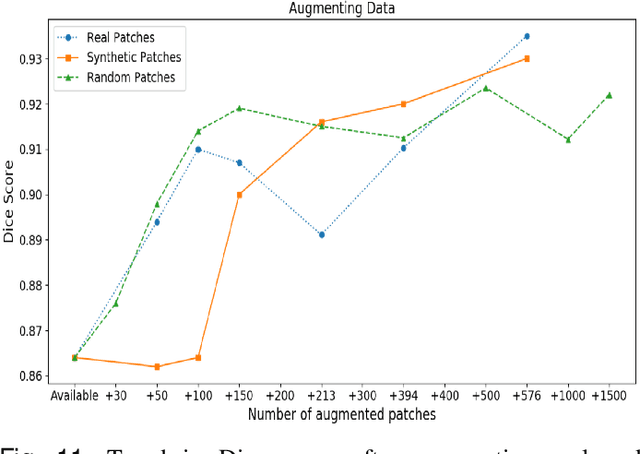

Synthetic images can be used for the development and evaluation of deep learning algorithms in the context of limited availability of annotations. In the field of computational pathology where histology images are large and visual context is crucial, synthesis of large tissue images via generative modeling is a challenging task due to memory and computing constraints hindering the generation of large images. To address this challenge, we propose a novel framework named as SAFRON to construct realistic large tissue image tiles from ground truth annotations while preserving morphological features and with minimal boundary artifacts at the seams. To this end, we train the proposed SAFRON framework based on conditional generative adversarial networks on large tissue image tiles from the Colorectal Adenocarcinoma Gland (CRAG) and DigestPath datasets. We demonstrate that our model can generate high quality and realistic image tiles of arbitrary large size after training it on relatively small image patches. We also show that training on synthetic data generated by SAFRON can significantly boost the performance of a standard algorithm for gland segmentation of colorectal cancer tissue images. Sample high resolution images generated using SAFRON are available at the URL:https://warwick.ac.uk/TIALab/SAFRON
Semantic Segmentation of Colon Glands with Deep Convolutional Neural Networks and Total Variation Segmentation
Oct 10, 2017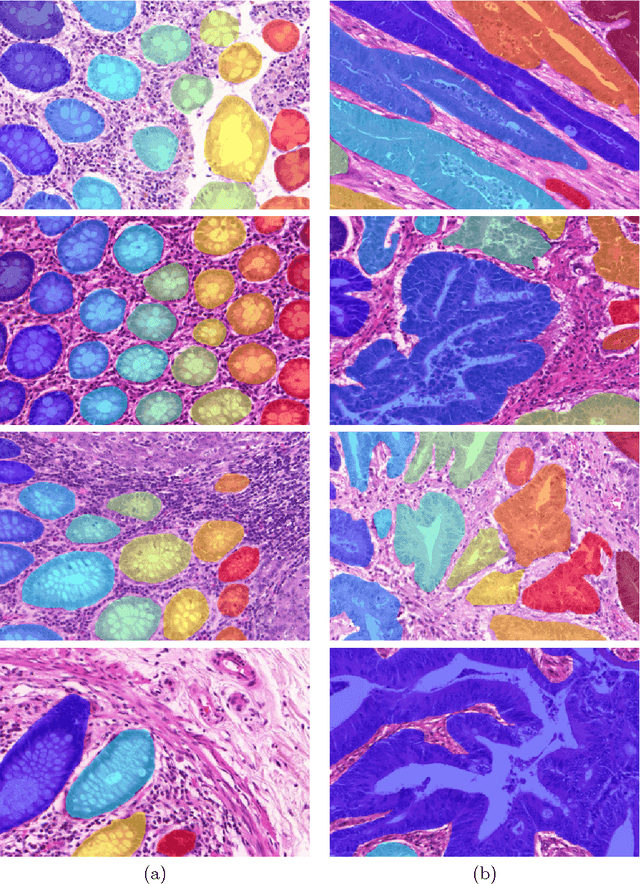
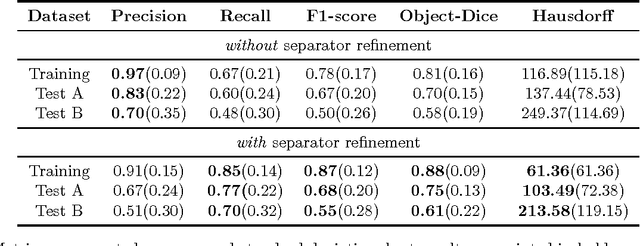
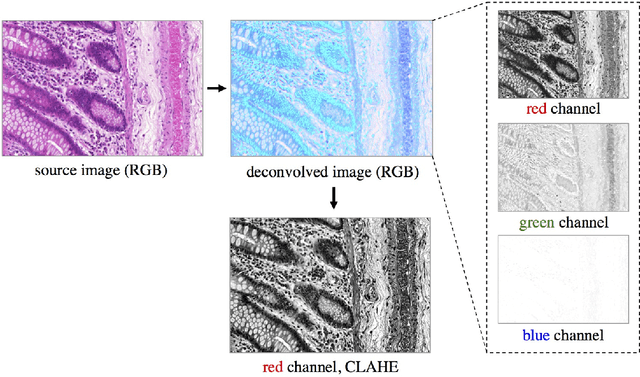
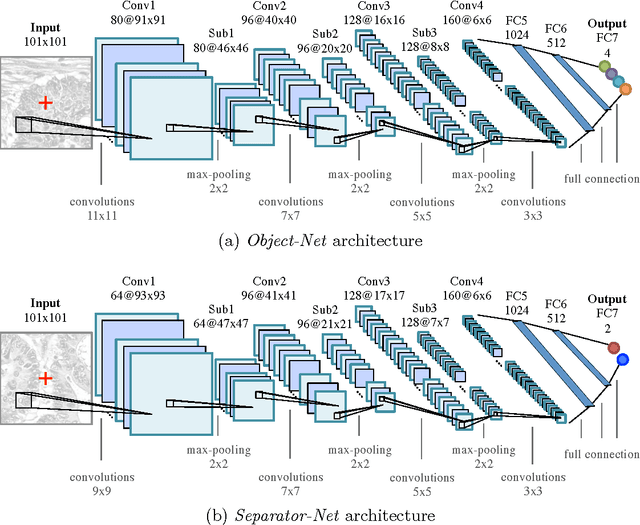
Segmentation of histopathology sections is an ubiquitous requirement in digital pathology and due to the large variability of biological tissue, machine learning techniques have shown superior performance over standard image processing methods. As part of the GlaS@MICCAI2015 colon gland segmentation challenge, we present a learning-based algorithm to segment glands in tissue of benign and malignant colorectal cancer. Images are preprocessed according to the Hematoxylin-Eosin staining protocol and two deep convolutional neural networks (CNN) are trained as pixel classifiers. The CNN predictions are then regularized using a figure-ground segmentation based on weighted total variation to produce the final segmentation result. On two test sets, our approach achieves a tissue classification accuracy of 98% and 94%, making use of the inherent capability of our system to distinguish between benign and malignant tissue.
Gland Segmentation in Colon Histology Images: The GlaS Challenge Contest
Sep 01, 2016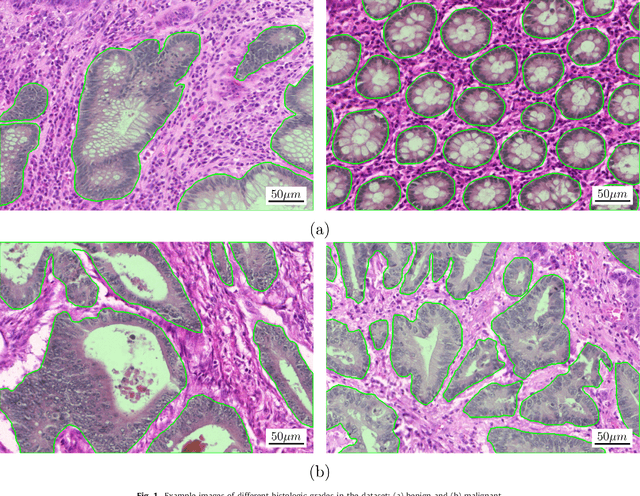
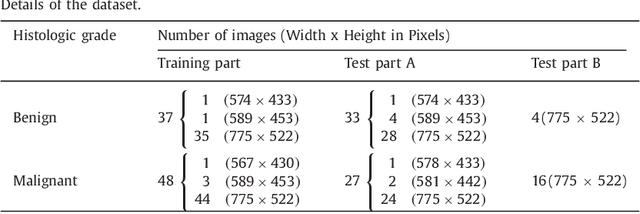
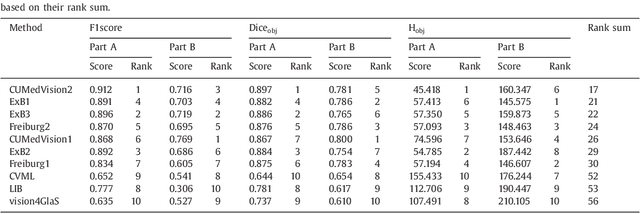
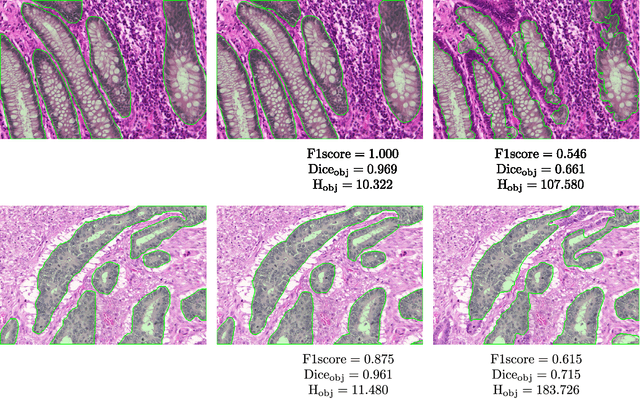
Colorectal adenocarcinoma originating in intestinal glandular structures is the most common form of colon cancer. In clinical practice, the morphology of intestinal glands, including architectural appearance and glandular formation, is used by pathologists to inform prognosis and plan the treatment of individual patients. However, achieving good inter-observer as well as intra-observer reproducibility of cancer grading is still a major challenge in modern pathology. An automated approach which quantifies the morphology of glands is a solution to the problem. This paper provides an overview to the Gland Segmentation in Colon Histology Images Challenge Contest (GlaS) held at MICCAI'2015. Details of the challenge, including organization, dataset and evaluation criteria, are presented, along with the method descriptions and evaluation results from the top performing methods.
MILD-Net: Minimal Information Loss Dilated Network for Gland Instance Segmentation in Colon Histology Images
Jun 07, 2018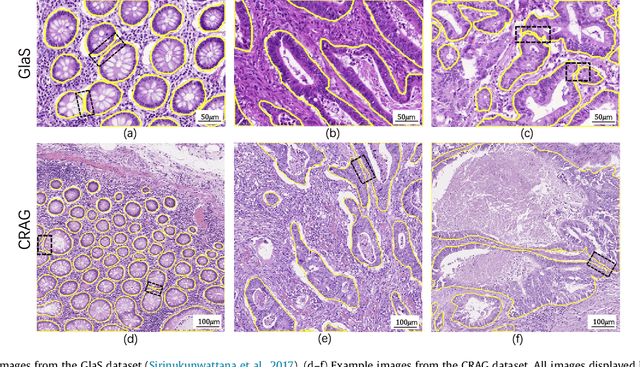
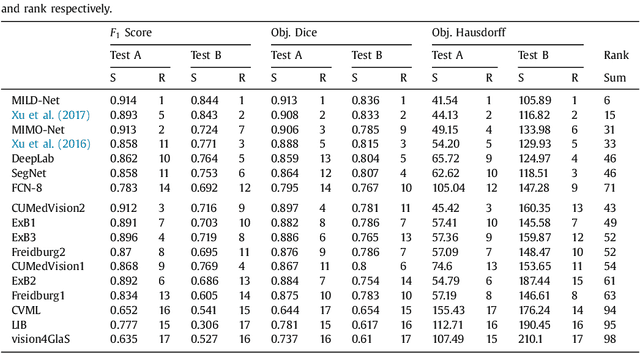
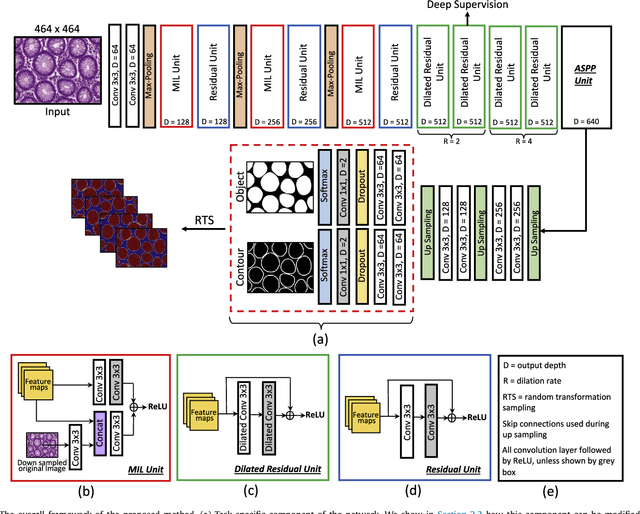
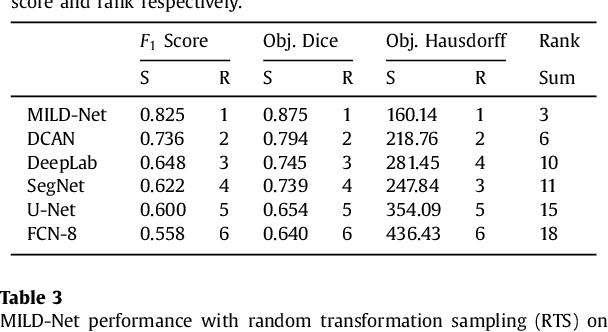
The analysis of glandular morphology within colon histopathology images is a crucial step in determining the stage of colon cancer. Despite the importance of this task, manual segmentation is laborious, time-consuming and can suffer from subjectivity among pathologists. The rise of computational pathology has led to the development of automated methods for gland segmentation that aim to overcome the challenges of manual segmentation. However, this task is non-trivial due to the large variability in glandular appearance and the difficulty in differentiating between certain glandular and non-glandular histological structures. Furthermore, within pathological practice, a measure of uncertainty is essential for diagnostic decision making. For example, ambiguous areas may require further examination from numerous pathologists. To address these challenges, we propose a fully convolutional neural network that counters the loss of information caused by max-pooling by re-introducing the original image at multiple points within the network. We also use atrous spatial pyramid pooling with varying dilation rates for resolution maintenance and multi-level aggregation. To incorporate uncertainty, we introduce random transformations during test time for an enhanced segmentation result that simultaneously generates an uncertainty map, highlighting areas of ambiguity. We show that this map can be used to define a metric for disregarding predictions with high uncertainty. The proposed network achieves state-of-the-art performance on the GlaS challenge dataset, as part of MICCAI 2015, and on a second independent colorectal adenocarcinoma dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge