Brain Lesion Segmentation From Mri
Papers and Code
Comparative evaluation of training strategies using partially labelled datasets for segmentation of white matter hyperintensities and stroke lesions in FLAIR MRI
Jan 28, 2026White matter hyperintensities (WMH) and ischaemic stroke lesions (ISL) are imaging features associated with cerebral small vessel disease (SVD) that are visible on brain magnetic resonance imaging (MRI) scans. The development and validation of deep learning models to segment and differentiate these features is difficult because they visually confound each other in the fluid-attenuated inversion recovery (FLAIR) sequence and often appear in the same subject. We investigated six strategies for training a combined WMH and ISL segmentation model using partially labelled data. We combined privately held fully and partially labelled datasets with publicly available partially labelled datasets to yield a total of 2052 MRI volumes, with 1341 and 1152 containing ground truth annotations for WMH and ISL respectively. We found that several methods were able to effectively leverage the partially labelled data to improve model performance, with the use of pseudolabels yielding the best result.
Towards Modality-Agnostic Continual Domain-Incremental Brain Lesion Segmentation
Jan 20, 2026Brain lesion segmentation from multi-modal MRI often assumes fixed modality sets or predefined pathologies, making existing models difficult to adapt across cohorts and imaging protocols. Continual learning (CL) offers a natural solution but current approaches either impose a maximum modality configuration or suffer from severe forgetting in buffer-free settings. We introduce CLMU-Net, a replay-based CL framework for 3D brain lesion segmentation that supports arbitrary and variable modality combinations without requiring prior knowledge of the maximum set. A conceptually simple yet effective channel-inflation strategy maps any modality subset into a unified multi-channel representation, enabling a single model to operate across diverse datasets. To enrich inherently local 3D patch features, we incorporate lightweight domain-conditioned textual embeddings that provide global modality-disease context for each training case. Forgetting is further reduced through principled replay using a compact buffer composed of both prototypical and challenging samples. Experiments on five heterogeneous MRI brain datasets demonstrate that CLMU-Net consistently outperforms popular CL baselines. Notably, our method yields an average Dice score improvement of $\geq$ 18\% while remaining robust under heterogeneous-modality conditions. These findings underscore the value of flexible modality handling, targeted replay, and global contextual cues for continual medical image segmentation. Our implementation is available at https://github.com/xmindflow/CLMU-Net.
POWDR: Pathology-preserving Outpainting with Wavelet Diffusion for 3D MRI
Jan 14, 2026Medical imaging datasets often suffer from class imbalance and limited availability of pathology-rich cases, which constrains the performance of machine learning models for segmentation, classification, and vision-language tasks. To address this challenge, we propose POWDR, a pathology-preserving outpainting framework for 3D MRI based on a conditioned wavelet diffusion model. Unlike conventional augmentation or unconditional synthesis, POWDR retains real pathological regions while generating anatomically plausible surrounding tissue, enabling diversity without fabricating lesions. Our approach leverages wavelet-domain conditioning to enhance high-frequency detail and mitigate blurring common in latent diffusion models. We introduce a random connected mask training strategy to overcome conditioning-induced collapse and improve diversity outside the lesion. POWDR is evaluated on brain MRI using BraTS datasets and extended to knee MRI to demonstrate tissue-agnostic applicability. Quantitative metrics (FID, SSIM, LPIPS) confirm image realism, while diversity analysis shows significant improvement with random-mask training (cosine similarity reduced from 0.9947 to 0.9580; KL divergence increased from 0.00026 to 0.01494). Clinically relevant assessments reveal gains in tumor segmentation performance using nnU-Net, with Dice scores improving from 0.6992 to 0.7137 when adding 50 synthetic cases. Tissue volume analysis indicates no significant differences for CSF and GM compared to real images. These findings highlight POWDR as a practical solution for addressing data scarcity and class imbalance in medical imaging. The method is extensible to multiple anatomies and offers a controllable framework for generating diverse, pathology-preserving synthetic data to support robust model development.
MGML: A Plug-and-Play Meta-Guided Multi-Modal Learning Framework for Incomplete Multimodal Brain Tumor Segmentation
Dec 30, 2025Leveraging multimodal information from Magnetic Resonance Imaging (MRI) plays a vital role in lesion segmentation, especially for brain tumors. However, in clinical practice, multimodal MRI data are often incomplete, making it challenging to fully utilize the available information. Therefore, maximizing the utilization of this incomplete multimodal information presents a crucial research challenge. We present a novel meta-guided multi-modal learning (MGML) framework that comprises two components: meta-parameterized adaptive modality fusion and consistency regularization module. The meta-parameterized adaptive modality fusion (Meta-AMF) enables the model to effectively integrate information from multiple modalities under varying input conditions. By generating adaptive soft-label supervision signals based on the available modalities, Meta-AMF explicitly promotes more coherent multimodal fusion. In addition, the consistency regularization module enhances segmentation performance and implicitly reinforces the robustness and generalization of the overall framework. Notably, our approach does not alter the original model architecture and can be conveniently integrated into the training pipeline for end-to-end model optimization. We conducted extensive experiments on the public BraTS2020 and BraTS2023 datasets. Compared to multiple state-of-the-art methods from previous years, our method achieved superior performance. On BraTS2020, for the average Dice scores across fifteen missing modality combinations, building upon the baseline, our method obtained scores of 87.55, 79.36, and 62.67 for the whole tumor (WT), the tumor core (TC), and the enhancing tumor (ET), respectively. We have made our source code publicly available at https://github.com/worldlikerr/MGML.
Adaptable Segmentation Pipeline for Diverse Brain Tumors with Radiomic-guided Subtyping and Lesion-Wise Model Ensemble
Dec 16, 2025
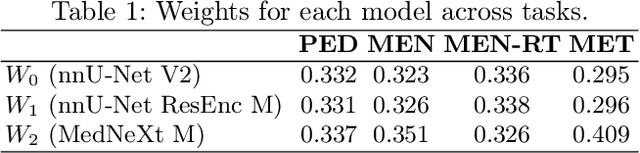
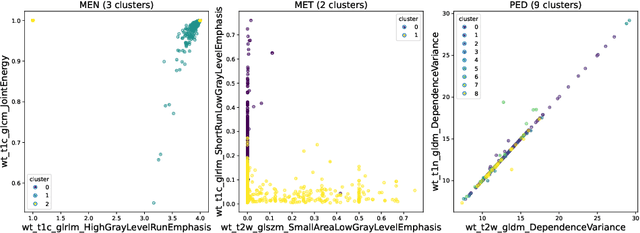
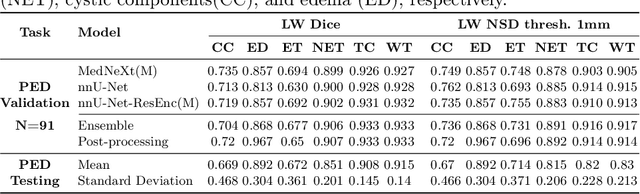
Robust and generalizable segmentation of brain tumors on multi-parametric magnetic resonance imaging (MRI) remains difficult because tumor types differ widely. The BraTS 2025 Lighthouse Challenge benchmarks segmentation methods on diverse high-quality datasets of adult and pediatric tumors: multi-consortium international pediatric brain tumor segmentation (PED), preoperative meningioma tumor segmentation (MEN), meningioma radiotherapy segmentation (MEN-RT), and segmentation of pre- and post-treatment brain metastases (MET). We present a flexible, modular, and adaptable pipeline that improves segmentation performance by selecting and combining state-of-the-art models and applying tumor- and lesion-specific processing before and after training. Radiomic features extracted from MRI help detect tumor subtype, ensuring a more balanced training. Custom lesion-level performance metrics determine the influence of each model in the ensemble and optimize post-processing that further refines the predictions, enabling the workflow to tailor every step to each case. On the BraTS testing sets, our pipeline achieved performance comparable to top-ranked algorithms across multiple challenges. These findings confirm that custom lesion-aware processing and model selection yield robust segmentations yet without locking the method to a specific network architecture. Our method has the potential for quantitative tumor measurement in clinical practice, supporting diagnosis and prognosis.
Large-scale modality-invariant foundation models for brain MRI analysis: Application to lesion segmentation
Nov 14, 2025The field of computer vision is undergoing a paradigm shift toward large-scale foundation model pre-training via self-supervised learning (SSL). Leveraging large volumes of unlabeled brain MRI data, such models can learn anatomical priors that improve few-shot performance in diverse neuroimaging tasks. However, most SSL frameworks are tailored to natural images, and their adaptation to capture multi-modal MRI information remains underexplored. This work proposes a modality-invariant representation learning setup and evaluates its effectiveness in stroke and epilepsy lesion segmentation, following large-scale pre-training. Experimental results suggest that despite successful cross-modality alignment, lesion segmentation primarily benefits from preserving fine-grained modality-specific features. Model checkpoints and code are made publicly available.
Segmentation of Ischemic Stroke Lesions using Transfer Learning on Multi-sequence MRI
Nov 10, 2025The accurate understanding of ischemic stroke lesions is critical for efficient therapy and prognosis of stroke patients. Magnetic resonance imaging (MRI) is sensitive to acute ischemic stroke and is a common diagnostic method for stroke. However, manual lesion segmentation performed by experts is tedious, time-consuming, and prone to observer inconsistency. Automatic medical image analysis methods have been proposed to overcome this challenge. However, previous approaches have relied on hand-crafted features that may not capture the irregular and physiologically complex shapes of ischemic stroke lesions. In this study, we present a novel framework for quickly and automatically segmenting ischemic stroke lesions on various MRI sequences, including T1-weighted, T2-weighted, DWI, and FLAIR. The proposed methodology is validated on the ISLES 2015 Brain Stroke sequence dataset, where we trained our model using the Res-Unet architecture twice: first, with pre-existing weights, and then without, to explore the benefits of transfer learning. Evaluation metrics, including the Dice score and sensitivity, were computed across 3D volumes. Finally, a Majority Voting Classifier was integrated to amalgamate the outcomes from each axis, resulting in a comprehensive segmentation method. Our efforts culminated in achieving a Dice score of 80.5\% and an accuracy of 74.03\%, showcasing the efficacy of our segmentation approach.
SYNAPSE-Net: A Unified Framework with Lesion-Aware Hierarchical Gating for Robust Segmentation of Heterogeneous Brain Lesions
Oct 30, 2025Automated segmentation of heterogeneous brain lesions from multi-modal MRI remains a critical challenge in clinical neuroimaging. Current deep learning models are typically specialized `point solutions' that lack generalization and high performance variance, limiting their clinical reliability. To address these gaps, we propose the Unified Multi-Stream SYNAPSE-Net, an adaptive framework designed for both generalization and robustness. The framework is built on a novel hybrid architecture integrating multi-stream CNN encoders, a Swin Transformer bottleneck for global context, a dynamic cross-modal attention fusion (CMAF) mechanism, and a hierarchical gated decoder for high-fidelity mask reconstruction. The architecture is trained with a variance reduction strategy that combines pathology specific data augmentation and difficulty-aware sampling method. The model was evaluated on three different challenging public datasets: the MICCAI 2017 WMH Challenge, the ISLES 2022 Challenge, and the BraTS 2020 Challenge. Our framework attained a state-of-the-art DSC value of 0.831 with the HD95 value of 3.03 in the WMH dataset. For ISLES 2022, it achieved the best boundary accuracy with a statistically significant difference (HD95 value of 9.69). For BraTS 2020, it reached the highest DSC value for the tumor core region (0.8651). These experimental findings suggest that our unified adaptive framework achieves state-of-the-art performance across multiple brain pathologies, providing a robust and clinically feasible solution for automated segmentation. The source code and the pre-trained models are available at https://github.com/mubid-01/SYNAPSE-Net-pre.
Modality-Agnostic Input Channels Enable Segmentation of Brain lesions in Multimodal MRI with Sequences Unavailable During Training
Sep 11, 2025Segmentation models are important tools for the detection and analysis of lesions in brain MRI. Depending on the type of brain pathology that is imaged, MRI scanners can acquire multiple, different image modalities (contrasts). Most segmentation models for multimodal brain MRI are restricted to fixed modalities and cannot effectively process new ones at inference. Some models generalize to unseen modalities but may lose discriminative modality-specific information. This work aims to develop a model that can perform inference on data that contain image modalities unseen during training, previously seen modalities, and heterogeneous combinations of both, thus allowing a user to utilize any available imaging modalities. We demonstrate this is possible with a simple, thus practical alteration to the U-net architecture, by integrating a modality-agnostic input channel or pathway, alongside modality-specific input channels. To train this modality-agnostic component, we develop an image augmentation scheme that synthesizes artificial MRI modalities. Augmentations differentially alter the appearance of pathological and healthy brain tissue to create artificial contrasts between them while maintaining realistic anatomical integrity. We evaluate the method using 8 MRI databases that include 5 types of pathologies (stroke, tumours, traumatic brain injury, multiple sclerosis and white matter hyperintensities) and 8 modalities (T1, T1+contrast, T2, PD, SWI, DWI, ADC and FLAIR). The results demonstrate that the approach preserves the ability to effectively process MRI modalities encountered during training, while being able to process new, unseen modalities to improve its segmentation. Project code: https://github.com/Anthony-P-Addison/AGN-MOD-SEG
AREPAS: Anomaly Detection in Fine-Grained Anatomy with Reconstruction-Based Semantic Patch-Scoring
Sep 16, 2025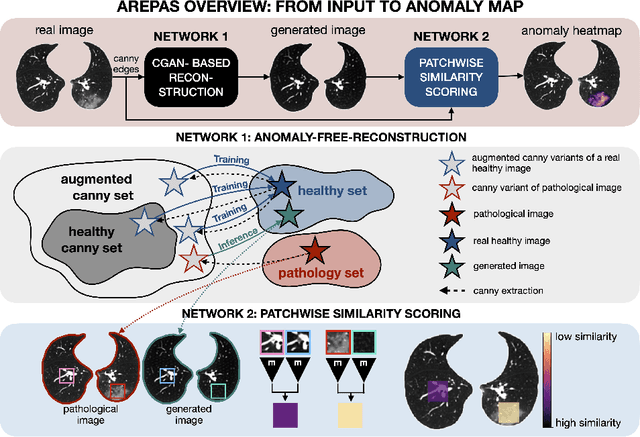
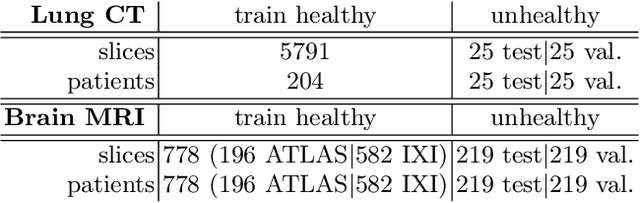
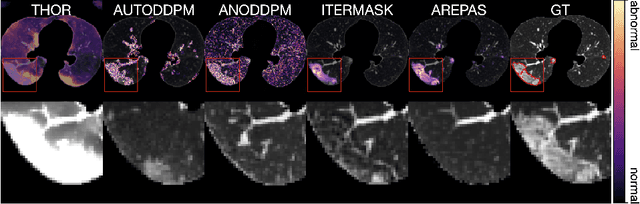
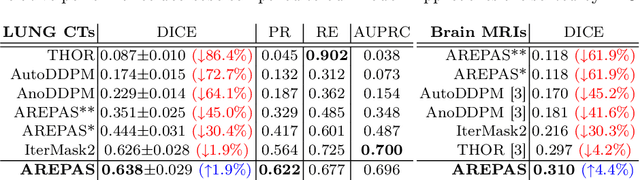
Early detection of newly emerging diseases, lesion severity assessment, differentiation of medical conditions and automated screening are examples for the wide applicability and importance of anomaly detection (AD) and unsupervised segmentation in medicine. Normal fine-grained tissue variability such as present in pulmonary anatomy is a major challenge for existing generative AD methods. Here, we propose a novel generative AD approach addressing this issue. It consists of an image-to-image translation for anomaly-free reconstruction and a subsequent patch similarity scoring between observed and generated image-pairs for precise anomaly localization. We validate the new method on chest computed tomography (CT) scans for the detection and segmentation of infectious disease lesions. To assess generalizability, we evaluate the method on an ischemic stroke lesion segmentation task in T1-weighted brain MRI. Results show improved pixel-level anomaly segmentation in both chest CTs and brain MRIs, with relative DICE score improvements of +1.9% and +4.4%, respectively, compared to other state-of-the-art reconstruction-based methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge