Ravi Soni
Synthetic Volumetric Data Generation Enables Zero-Shot Generalization of Foundation Models in 3D Medical Image Segmentation
Jan 18, 2026Abstract:Foundation models such as Segment Anything Model 2 (SAM 2) exhibit strong generalization on natural images and videos but perform poorly on medical data due to differences in appearance statistics, imaging physics, and three-dimensional structure. To address this gap, we introduce SynthFM-3D, an analytical framework that mathematically models 3D variability in anatomy, contrast, boundary definition, and noise to generate synthetic data for training promptable segmentation models without real annotations. We fine-tuned SAM 2 on 10,000 SynthFM-3D volumes and evaluated it on eleven anatomical structures across three medical imaging modalities (CT, MR, ultrasound) from five public datasets. SynthFM-3D training led to consistent and statistically significant Dice score improvements over the pretrained SAM 2 baseline, demonstrating stronger zero-shot generalization across modalities. When compared with the supervised SAM-Med3D model on unseen cardiac ultrasound data, SynthFM-3D achieved 2-3x higher Dice scores, establishing analytical 3D data modeling as an effective pathway to modality-agnostic medical segmentation.
POWDR: Pathology-preserving Outpainting with Wavelet Diffusion for 3D MRI
Jan 14, 2026Abstract:Medical imaging datasets often suffer from class imbalance and limited availability of pathology-rich cases, which constrains the performance of machine learning models for segmentation, classification, and vision-language tasks. To address this challenge, we propose POWDR, a pathology-preserving outpainting framework for 3D MRI based on a conditioned wavelet diffusion model. Unlike conventional augmentation or unconditional synthesis, POWDR retains real pathological regions while generating anatomically plausible surrounding tissue, enabling diversity without fabricating lesions. Our approach leverages wavelet-domain conditioning to enhance high-frequency detail and mitigate blurring common in latent diffusion models. We introduce a random connected mask training strategy to overcome conditioning-induced collapse and improve diversity outside the lesion. POWDR is evaluated on brain MRI using BraTS datasets and extended to knee MRI to demonstrate tissue-agnostic applicability. Quantitative metrics (FID, SSIM, LPIPS) confirm image realism, while diversity analysis shows significant improvement with random-mask training (cosine similarity reduced from 0.9947 to 0.9580; KL divergence increased from 0.00026 to 0.01494). Clinically relevant assessments reveal gains in tumor segmentation performance using nnU-Net, with Dice scores improving from 0.6992 to 0.7137 when adding 50 synthetic cases. Tissue volume analysis indicates no significant differences for CSF and GM compared to real images. These findings highlight POWDR as a practical solution for addressing data scarcity and class imbalance in medical imaging. The method is extensible to multiple anatomies and offers a controllable framework for generating diverse, pathology-preserving synthetic data to support robust model development.
Towards Better Ultrasound Video Segmentation Foundation Model: An Empirical study on SAM2 Finetuning from Data Perspective
Nov 07, 2025



Abstract:Ultrasound (US) video segmentation remains a challenging problem due to strong inter- and intra-dataset variability, motion artifacts, and limited annotated data. Although foundation models such as Segment Anything Model 2 (SAM2) demonstrate strong zero-shot and prompt-guided segmentation capabilities, their performance deteriorates substantially when transferred to medical imaging domains. Current adaptation studies mainly emphasize architectural modifications, while the influence of data characteristics and training regimes has not been systematically examined. In this study, we present a comprehensive, data-centric investigation of SAM2 adaptation for ultrasound video segmentation. We analyze how training-set size, video duration, and augmentation schemes affect adaptation performance under three paradigms: task-specific fine-tuning, intermediate adaptation, and multi-task joint training, across five SAM2 variants and multiple prompting modes. We further design six ultrasound-specific augmentations, assessing their effect relative to generic strategies. Experiments on three representative ultrasound datasets reveal that data scale and temporal context play a more decisive role than model architecture or initialization. Moreover, joint training offers an efficient compromise between modality alignment and task specialization. This work aims to provide empirical insights for developing efficient, data-aware adaptation pipelines for SAM2 in ultrasound video analysis.
SynthFM: Training Modality-agnostic Foundation Models for Medical Image Segmentation without Real Medical Data
Apr 11, 2025



Abstract:Foundation models like the Segment Anything Model (SAM) excel in zero-shot segmentation for natural images but struggle with medical image segmentation due to differences in texture, contrast, and noise. Annotating medical images is costly and requires domain expertise, limiting large-scale annotated data availability. To address this, we propose SynthFM, a synthetic data generation framework that mimics the complexities of medical images, enabling foundation models to adapt without real medical data. Using SAM's pretrained encoder and training the decoder from scratch on SynthFM's dataset, we evaluated our method on 11 anatomical structures across 9 datasets (CT, MRI, and Ultrasound). SynthFM outperformed zero-shot baselines like SAM and MedSAM, achieving superior results under different prompt settings and on out-of-distribution datasets.
SAS: Segment Anything Small for Ultrasound -- A Non-Generative Data Augmentation Technique for Robust Deep Learning in Ultrasound Imaging
Mar 07, 2025



Abstract:Accurate segmentation of anatomical structures in ultrasound (US) images, particularly small ones, is challenging due to noise and variability in imaging conditions (e.g., probe position, patient anatomy, tissue characteristics and pathology). To address this, we introduce Segment Anything Small (SAS), a simple yet effective scale- and texture-aware data augmentation technique designed to enhance the performance of deep learning models for segmenting small anatomical structures in ultrasound images. SAS employs a dual transformation strategy: (1) simulating diverse organ scales by resizing and embedding organ thumbnails into a black background, and (2) injecting noise into regions of interest to simulate varying tissue textures. These transformations generate realistic and diverse training data without introducing hallucinations or artifacts, improving the model's robustness to noise and variability. We fine-tuned a promptable foundation model on a controlled organ-specific medical imaging dataset and evaluated its performance on one internal and five external datasets. Experimental results demonstrate significant improvements in segmentation performance, with Dice score gains of up to 0.35 and an average improvement of 0.16 [95% CI 0.132,0.188]. Additionally, our iterative point prompts provide precise control and adaptive refinement, achieving performance comparable to bounding box prompts with just two points. SAS enhances model robustness and generalizability across diverse anatomical structures and imaging conditions, particularly for small structures, without compromising the accuracy of larger ones. By offering a computationally efficient solution that eliminates the need for extensive human labeling efforts, SAS emerges as a powerful tool for advancing medical image analysis, particularly in resource-constrained settings.
Is SAM 2 Better than SAM in Medical Image Segmentation?
Aug 08, 2024Abstract:Segment Anything Model (SAM) demonstrated impressive performance in zero-shot promptable segmentation on natural images. The recently released Segment Anything Model 2 (SAM 2) model claims to have better performance than SAM on images while extending the model's capabilities to video segmentation. It is important to evaluate the recent model's ability in medical image segmentation in a zero-shot promptable manner. In this work, we performed extensive studies with multiple datasets from different imaging modalities to compare the performance between SAM and SAM 2. We used two point prompt strategies: (i) single positive prompt near the centroid of the target structure and (ii) additional positive prompts placed randomly within the target structure. The evaluation included 21 unique organ-modality combinations including abdominal structures, cardiac structures, and fetal head images acquired from publicly available MRI, CT, and Ultrasound datasets. The preliminary results, based on 2D images, indicate that while SAM 2 may perform slightly better in a few cases, but it does not in general surpass SAM for medical image segmentation. Especially when the contrast is lower like in CT, Ultrasound images, SAM 2 performs poorly than SAM. For MRI images, SAM 2 performs at par or better than SAM. Similar to SAM, SAM 2 also suffers from over-segmentation issue especially when the boundaries of the to-be-segmented organ is fuzzy in nature.
Adversarial Focal Loss: Asking Your Discriminator for Hard Examples
Jul 15, 2022
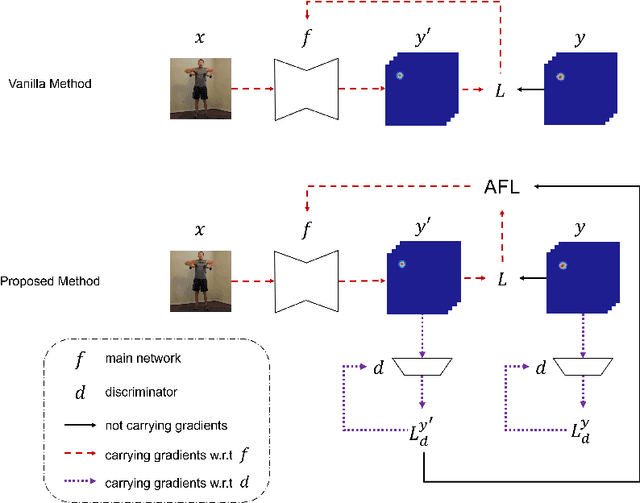
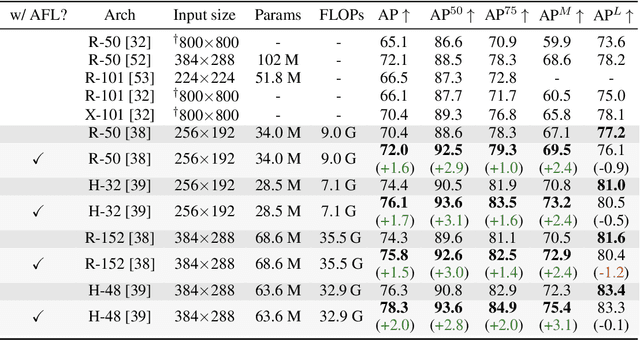
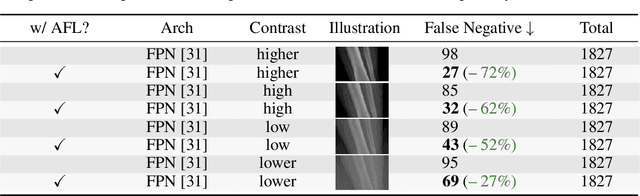
Abstract:Focal Loss has reached incredible popularity as it uses a simple technique to identify and utilize hard examples to achieve better performance on classification. However, this method does not easily generalize outside of classification tasks, such as in keypoint detection. In this paper, we propose a novel adaptation of Focal Loss for keypoint detection tasks, called Adversarial Focal Loss (AFL). AFL not only is semantically analogous to Focal loss, but also works as a plug-and-chug upgrade for arbitrary loss functions. While Focal Loss requires output from a classifier, AFL leverages a separate adversarial network to produce a difficulty score for each input. This difficulty score can then be used to dynamically prioritize learning on hard examples, even in absence of a classifier. In this work, we show AFL's effectiveness in enhancing existing methods in keypoint detection and verify its capability to re-weigh examples based on difficulty.
Pristine annotations-based multi-modal trained artificial intelligence solution to triage chest X-ray for COVID-19
Nov 10, 2020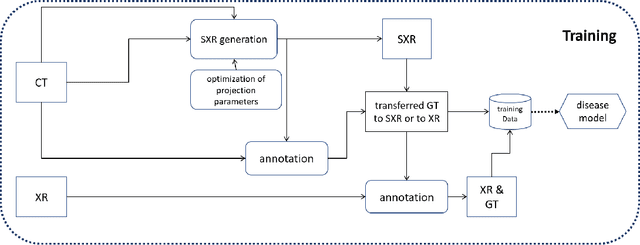
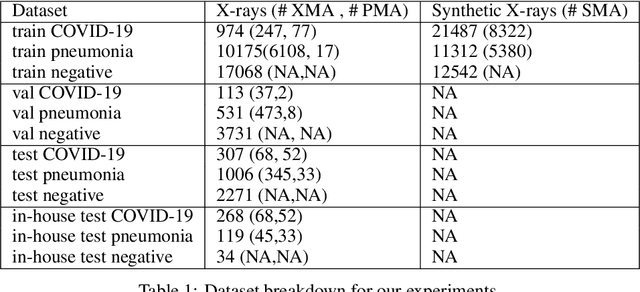
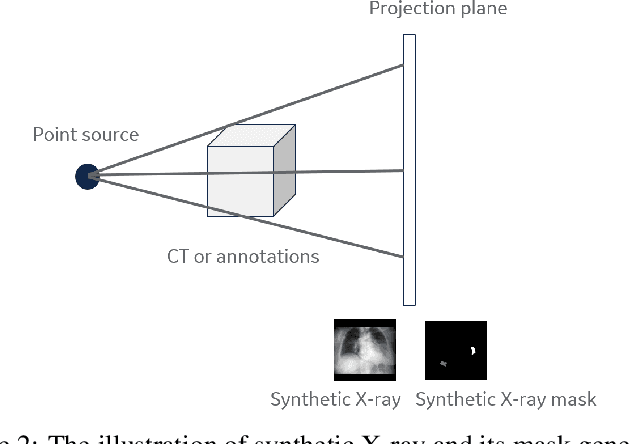
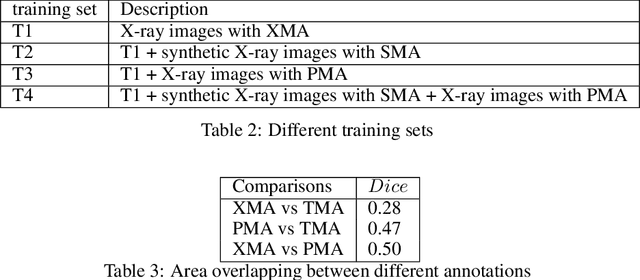
Abstract:The COVID-19 pandemic continues to spread and impact the well-being of the global population. The front-line modalities including computed tomography (CT) and X-ray play an important role for triaging COVID patients. Considering the limited access of resources (both hardware and trained personnel) and decontamination considerations, CT may not be ideal for triaging suspected subjects. Artificial intelligence (AI) assisted X-ray based applications for triaging and monitoring require experienced radiologists to identify COVID patients in a timely manner and to further delineate the disease region boundary are seen as a promising solution. Our proposed solution differs from existing solutions by industry and academic communities, and demonstrates a functional AI model to triage by inferencing using a single x-ray image, while the deep-learning model is trained using both X-ray and CT data. We report on how such a multi-modal training improves the solution compared to X-ray only training. The multi-modal solution increases the AUC (area under the receiver operating characteristic curve) from 0.89 to 0.93 and also positively impacts the Dice coefficient (0.59 to 0.62) for localizing the pathology. To the best our knowledge, it is the first X-ray solution by leveraging multi-modal information for the development.
Transformer Based Language Models for Similar Text Retrieval and Ranking
May 21, 2020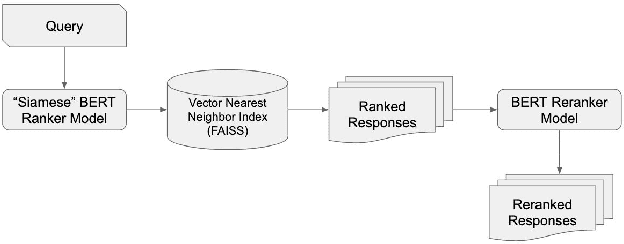
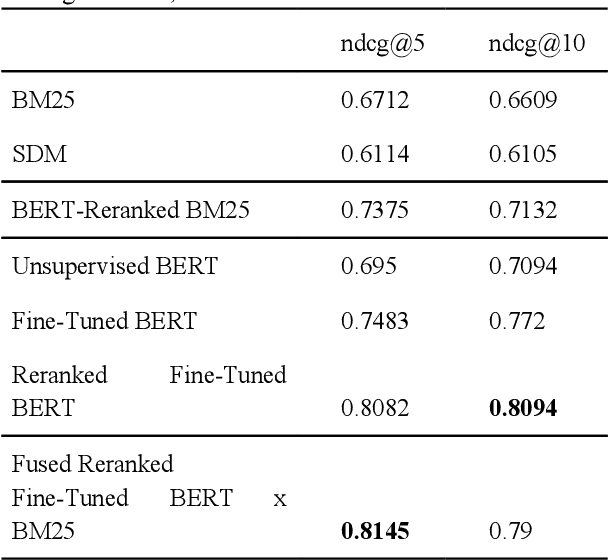
Abstract:Most approaches for similar text retrieval and ranking with long natural language queries rely at some level on queries and responses having words in common with each other. Recent applications of transformer-based neural language models to text retrieval and ranking problems have been very promising, but still involve a two-step process in which result candidates are first obtained through bag-of-words-based approaches, and then reranked by a neural transformer. In this paper, we introduce novel approaches for effectively applying neural transformer models to similar text retrieval and ranking without an initial bag-of-words-based step. By eliminating the bag-of-words-based step, our approach is able to accurately retrieve and rank results even when they have no non-stopwords in common with the query. We accomplish this by using bidirectional encoder representations from transformers (BERT) to create vectorized representations of sentence-length texts, along with a vector nearest neighbor search index. We demonstrate both supervised and unsupervised means of using BERT to accomplish this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge