Nishad Kulkarni
Improving Pre-trained Segmentation Models using Post-Processing
Dec 16, 2025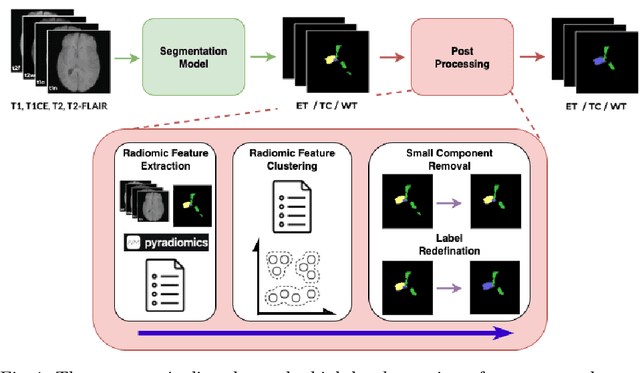
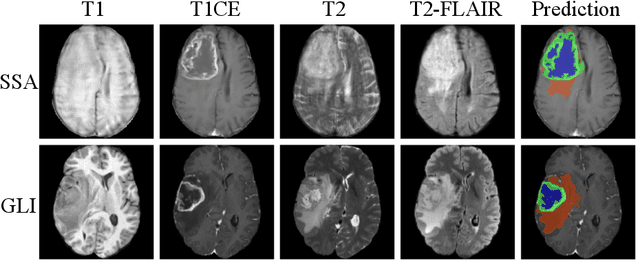

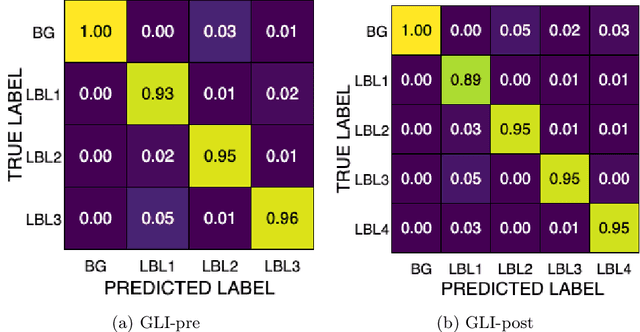
Abstract:Gliomas are the most common malignant brain tumors in adults and are among the most lethal. Despite aggressive treatment, the median survival rate is less than 15 months. Accurate multiparametric MRI (mpMRI) tumor segmentation is critical for surgical planning, radiotherapy, and disease monitoring. While deep learning models have improved the accuracy of automated segmentation, large-scale pre-trained models generalize poorly and often underperform, producing systematic errors such as false positives, label swaps, and slice discontinuities in slices. These limitations are further compounded by unequal access to GPU resources and the growing environmental cost of large-scale model training. In this work, we propose adaptive post-processing techniques to refine the quality of glioma segmentations produced by large-scale pretrained models developed for various types of tumors. We demonstrated the techniques in multiple BraTS 2025 segmentation challenge tasks, with the ranking metric improving by 14.9 % for the sub-Saharan Africa challenge and 0.9% for the adult glioma challenge. This approach promotes a shift in brain tumor segmentation research from increasingly complex model architectures to efficient, clinically aligned post-processing strategies that are precise, computationally fair, and sustainable.
Adaptable Segmentation Pipeline for Diverse Brain Tumors with Radiomic-guided Subtyping and Lesion-Wise Model Ensemble
Dec 16, 2025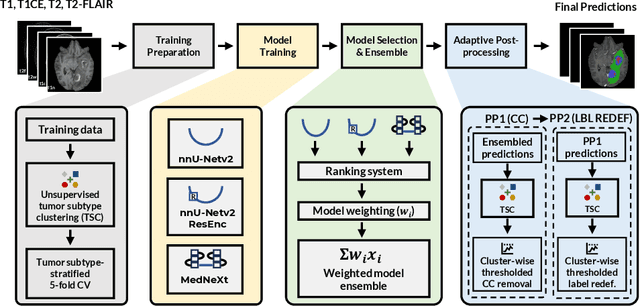
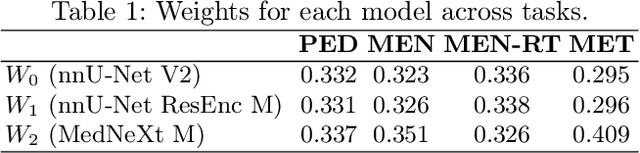
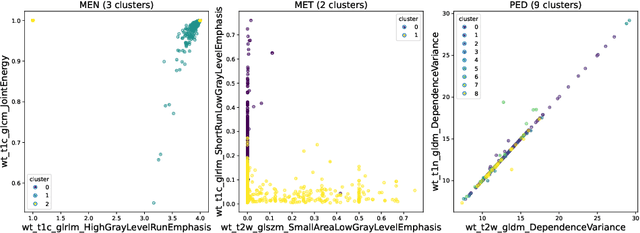
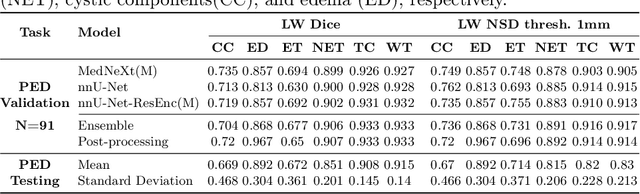
Abstract:Robust and generalizable segmentation of brain tumors on multi-parametric magnetic resonance imaging (MRI) remains difficult because tumor types differ widely. The BraTS 2025 Lighthouse Challenge benchmarks segmentation methods on diverse high-quality datasets of adult and pediatric tumors: multi-consortium international pediatric brain tumor segmentation (PED), preoperative meningioma tumor segmentation (MEN), meningioma radiotherapy segmentation (MEN-RT), and segmentation of pre- and post-treatment brain metastases (MET). We present a flexible, modular, and adaptable pipeline that improves segmentation performance by selecting and combining state-of-the-art models and applying tumor- and lesion-specific processing before and after training. Radiomic features extracted from MRI help detect tumor subtype, ensuring a more balanced training. Custom lesion-level performance metrics determine the influence of each model in the ensemble and optimize post-processing that further refines the predictions, enabling the workflow to tailor every step to each case. On the BraTS testing sets, our pipeline achieved performance comparable to top-ranked algorithms across multiple challenges. These findings confirm that custom lesion-aware processing and model selection yield robust segmentations yet without locking the method to a specific network architecture. Our method has the potential for quantitative tumor measurement in clinical practice, supporting diagnosis and prognosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge