Yingxu Wang
USBD: Universal Structural Basis Distillation for Source-Free Graph Domain Adaptation
Feb 09, 2026Abstract:SF-GDA is pivotal for privacy-preserving knowledge transfer across graph datasets. Although recent works incorporate structural information, they implicitly condition adaptation on the smoothness priors of sourcetrained GNNs, thereby limiting their generalization to structurally distinct targets. This dependency becomes a critical bottleneck under significant topological shifts, where the source model misinterprets distinct topological patterns unseen in the source domain as noise, rendering pseudo-label-based adaptation unreliable. To overcome this limitation, we propose the Universal Structural Basis Distillation, a framework that shifts the paradigm from adapting a biased model to learning a universal structural basis for SF-GDA. Instead of adapting a biased source model to a specific target, our core idea is to construct a structure-agnostic basis that proactively covers the full spectrum of potential topological patterns. Specifically, USBD employs a bi-level optimization framework to distill the source dataset into a compact structural basis. By enforcing the prototypes to span the full Dirichlet energy spectrum, the learned basis explicitly captures diverse topological motifs, ranging from low-frequency clusters to high-frequency chains, beyond those present in the source. This ensures that the learned basis creates a comprehensive structural covering capable of handling targets with disparate structures. For inference, we introduce a spectral-aware ensemble mechanism that dynamically activates the optimal prototype combination based on the spectral fingerprint of the target graph. Extensive experiments on benchmarks demonstrate that USBD significantly outperforms state-of-the-art methods, particularly in scenarios with severe structural shifts, while achieving superior computational efficiency by decoupling the adaptation cost from the target data scale.
Riemannian Flow Matching for Disentangled Graph Domain Adaptation
Jan 31, 2026Abstract:Graph Domain Adaptation (GDA) typically uses adversarial learning to align graph embeddings in Euclidean space. However, this paradigm suffers from two critical challenges: Structural Degeneration, where hierarchical and semantic representations are entangled, and Optimization Instability, which arises from oscillatory dynamics of minimax adversarial training. To tackle these issues, we propose DisRFM, a geometry-aware GDA framework that unifies Riemannian embedding and flow-based transport. First, to overcome structural degeneration, we embed graphs into a Riemannian manifold. By adopting polar coordinates, we explicitly disentangle structure (radius) from semantics (angle). Then, we enforce topology preservation through radial Wasserstein alignment and semantic discrimination via angular clustering, thereby preventing feature entanglement and collapse. Second, we address the instability of adversarial alignment by using Riemannian flow matching. This method learns a smooth vector field to guide source features toward the target along geodesic paths, guaranteeing stable convergence. The geometric constraints further guide the flow to maintain the disentangled structure during transport. Theoretically, we prove the asymptotic stability of the flow matching and derive a tighter bound for the target risk. Extensive experiments demonstrate that DisRFM consistently outperforms state-of-the-art methods.
A Comprehensive Survey of Self-Evolving AI Agents: A New Paradigm Bridging Foundation Models and Lifelong Agentic Systems
Aug 10, 2025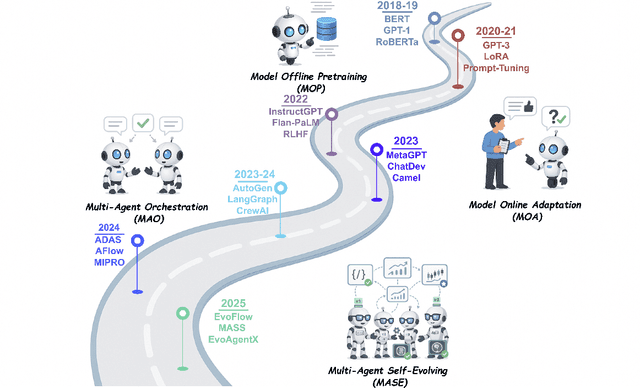
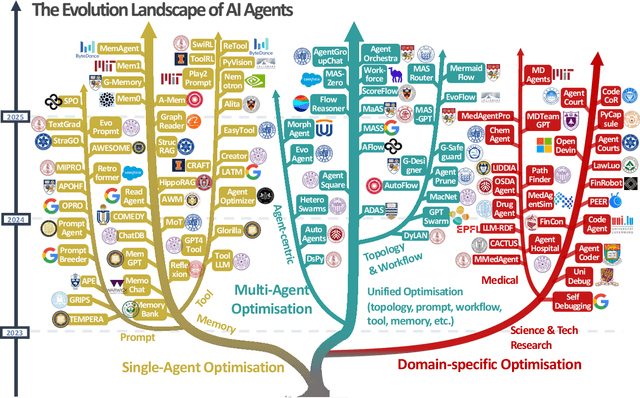
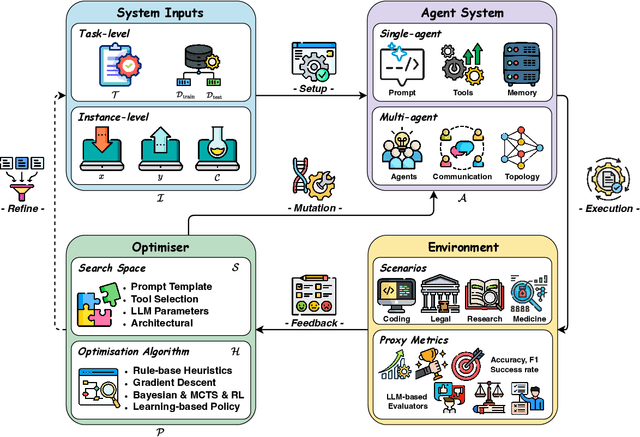
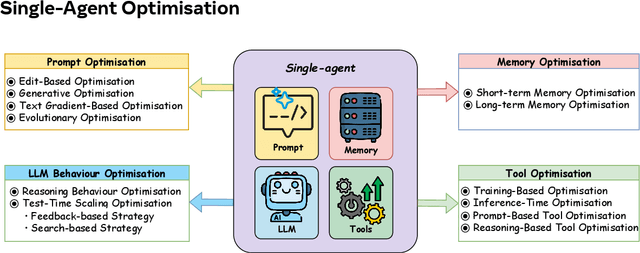
Abstract:Recent advances in large language models have sparked growing interest in AI agents capable of solving complex, real-world tasks. However, most existing agent systems rely on manually crafted configurations that remain static after deployment, limiting their ability to adapt to dynamic and evolving environments. To this end, recent research has explored agent evolution techniques that aim to automatically enhance agent systems based on interaction data and environmental feedback. This emerging direction lays the foundation for self-evolving AI agents, which bridge the static capabilities of foundation models with the continuous adaptability required by lifelong agentic systems. In this survey, we provide a comprehensive review of existing techniques for self-evolving agentic systems. Specifically, we first introduce a unified conceptual framework that abstracts the feedback loop underlying the design of self-evolving agentic systems. The framework highlights four key components: System Inputs, Agent System, Environment, and Optimisers, serving as a foundation for understanding and comparing different strategies. Based on this framework, we systematically review a wide range of self-evolving techniques that target different components of the agent system. We also investigate domain-specific evolution strategies developed for specialised fields such as biomedicine, programming, and finance, where optimisation objectives are tightly coupled with domain constraints. In addition, we provide a dedicated discussion on the evaluation, safety, and ethical considerations for self-evolving agentic systems, which are critical to ensuring their effectiveness and reliability. This survey aims to provide researchers and practitioners with a systematic understanding of self-evolving AI agents, laying the foundation for the development of more adaptive, autonomous, and lifelong agentic systems.
Nested Graph Pseudo-Label Refinement for Noisy Label Domain Adaptation Learning
Aug 01, 2025Abstract:Graph Domain Adaptation (GDA) facilitates knowledge transfer from labeled source graphs to unlabeled target graphs by learning domain-invariant representations, which is essential in applications such as molecular property prediction and social network analysis. However, most existing GDA methods rely on the assumption of clean source labels, which rarely holds in real-world scenarios where annotation noise is pervasive. This label noise severely impairs feature alignment and degrades adaptation performance under domain shifts. To address this challenge, we propose Nested Graph Pseudo-Label Refinement (NeGPR), a novel framework tailored for graph-level domain adaptation with noisy labels. NeGPR first pretrains dual branches, i.e., semantic and topology branches, by enforcing neighborhood consistency in the feature space, thereby reducing the influence of noisy supervision. To bridge domain gaps, NeGPR employs a nested refinement mechanism in which one branch selects high-confidence target samples to guide the adaptation of the other, enabling progressive cross-domain learning. Furthermore, since pseudo-labels may still contain noise and the pre-trained branches are already overfitted to the noisy labels in the source domain, NeGPR incorporates a noise-aware regularization strategy. This regularization is theoretically proven to mitigate the adverse effects of pseudo-label noise, even under the presence of source overfitting, thus enhancing the robustness of the adaptation process. Extensive experiments on benchmark datasets demonstrate that NeGPR consistently outperforms state-of-the-art methods under severe label noise, achieving gains of up to 12.7% in accuracy.
Dynamically Adaptive Reasoning via LLM-Guided MCTS for Efficient and Context-Aware KGQA
Aug 01, 2025Abstract:Knowledge Graph Question Answering (KGQA) aims to interpret natural language queries and perform structured reasoning over knowledge graphs by leveraging their relational and semantic structures to retrieve accurate answers. Recent KGQA methods primarily follow either retrieve-then-reason paradigm, relying on GNNs or heuristic rules for static paths extraction, or dynamic path generation strategies that use large language models (LLMs) with prompting to jointly perform retrieval and reasoning. However, the former suffers from limited adaptability due to static path extraction and lack of contextual refinement, while the latter incurs high computational costs and struggles with accurate path evaluation due to reliance on fixed scoring functions and extensive LLM calls. To address these issues, this paper proposes Dynamically Adaptive MCTS-based Reasoning (DAMR), a novel framework that integrates symbolic search with adaptive path evaluation for efficient and context-aware KGQA. DAMR employs a Monte Carlo Tree Search (MCTS) backbone guided by an LLM-based planner, which selects top-$k$ relevant relations at each step to reduce search space. To improve path evaluation accuracy, we introduce a lightweight Transformer-based scorer that performs context-aware plausibility estimation by jointly encoding the question and relation sequence through cross-attention, enabling the model to capture fine-grained semantic shifts during multi-hop reasoning. Furthermore, to alleviate the scarcity of high-quality supervision, DAMR incorporates a dynamic pseudo-path refinement mechanism that periodically generates training signals from partial paths explored during search, allowing the scorer to continuously adapt to the evolving distribution of reasoning trajectories. Extensive experiments on multiple KGQA benchmarks show that DAMR significantly outperforms state-of-the-art methods.
SEW: Self-Evolving Agentic Workflows for Automated Code Generation
May 24, 2025Abstract:Large Language Models (LLMs) have demonstrated effectiveness in code generation tasks. To enable LLMs to address more complex coding challenges, existing research has focused on crafting multi-agent systems with agentic workflows, where complex coding tasks are decomposed into sub-tasks, assigned to specialized agents. Despite their effectiveness, current approaches heavily rely on hand-crafted agentic workflows, with both agent topologies and prompts manually designed, which limits their ability to automatically adapt to different types of coding problems. To address these limitations and enable automated workflow design, we propose \textbf{S}elf-\textbf{E}volving \textbf{W}orkflow (\textbf{SEW}), a novel self-evolving framework that automatically generates and optimises multi-agent workflows. Extensive experiments on three coding benchmark datasets, including the challenging LiveCodeBench, demonstrate that our SEW can automatically design agentic workflows and optimise them through self-evolution, bringing up to 33\% improvement on LiveCodeBench compared to using the backbone LLM only. Furthermore, by investigating different representation schemes of workflow, we provide insights into the optimal way to encode workflow information with text.
Cross-Attention Graph Neural Networks for Inferring Gene Regulatory Networks with Skewed Degree Distribution
Dec 24, 2024Abstract:Inferencing Gene Regulatory Networks (GRNs) from gene expression data is a pivotal challenge in systems biology, and several innovative computational methods have been introduced. However, most of these studies have not considered the skewed degree distribution of genes. Specifically, some genes may regulate multiple target genes while some genes may be regulated by multiple regulator genes. Such a skewed degree distribution issue significantly complicates the application of directed graph embedding methods. To tackle this issue, we propose the Cross-Attention Complex Dual Graph Embedding Model (XATGRN). Our XATGRN employs a cross-attention mechanism to effectively capture intricate gene interactions from gene expression profiles. Additionally, it uses a Dual Complex Graph Embedding approach to manage the skewed degree distribution, thereby ensuring precise prediction of regulatory relationships and their directionality. Our model consistently outperforms existing state-of-the-art methods across various datasets, underscoring its efficacy in elucidating complex gene regulatory mechanisms. Our codes used in this paper are publicly available at: https://github.com/kikixiong/XATGRN.
SGAC: A Graph Neural Network Framework for Imbalanced and Structure-Aware AMP Classification
Dec 20, 2024Abstract:Classifying antimicrobial peptides(AMPs) from the vast array of peptides mined from metagenomic sequencing data is a significant approach to addressing the issue of antibiotic resistance. However, current AMP classification methods, primarily relying on sequence-based data, neglect the spatial structure of peptides, thereby limiting the accurate classification of AMPs. Additionally, the number of known AMPs is significantly lower than that of non-AMPs, leading to imbalanced datasets that reduce predictive accuracy for AMPs. To alleviate these two limitations, we first employ Omegafold to predict the three-dimensional spatial structures of AMPs and non-AMPs, constructing peptide graphs based on the amino acids' C$_\alpha$ positions. Building upon this, we propose a novel classification model named Spatial GNN-based AMP Classifier (SGAC). Our SGAC model employs a graph encoder based on Graph Neural Networks (GNNs) to process peptide graphs, generating high-dimensional representations that capture essential features from the three-dimensional spatial structure of amino acids. Then, to address the inherent imbalanced datasets, SGAC first incorporates Weight-enhanced Contrastive Learning, which clusters similar peptides while ensuring separation between dissimilar ones, using weighted contributions to emphasize AMP-specific features. Furthermore, SGAC employs Weight-enhanced Pseudo-label Distillation to dynamically generate high-confidence pseudo labels for ambiguous peptides, further refining predictions and promoting balanced learning between AMPs and non-AMPs. Experiments on publicly available AMP and non-AMP datasets demonstrate that SGAC significantly outperforms traditional sequence-based methods and achieves state-of-the-art performance among graph-based models, validating its effectiveness in AMP classification.
A Decade of Deep Learning: A Survey on The Magnificent Seven
Dec 13, 2024Abstract:Deep learning has fundamentally reshaped the landscape of artificial intelligence over the past decade, enabling remarkable achievements across diverse domains. At the heart of these developments lie multi-layered neural network architectures that excel at automatic feature extraction, leading to significant improvements in machine learning tasks. To demystify these advances and offer accessible guidance, we present a comprehensive overview of the most influential deep learning algorithms selected through a broad-based survey of the field. Our discussion centers on pivotal architectures, including Residual Networks, Transformers, Generative Adversarial Networks, Variational Autoencoders, Graph Neural Networks, Contrastive Language-Image Pre-training, and Diffusion models. We detail their historical context, highlight their mathematical foundations and algorithmic principles, and examine subsequent variants, extensions, and practical considerations such as training methodologies, normalization techniques, and learning rate schedules. Beyond historical and technical insights, we also address their applications, challenges, and potential research directions. This survey aims to serve as a practical manual for both newcomers seeking an entry point into cutting-edge deep learning methods and experienced researchers transitioning into this rapidly evolving domain.
DuSEGO: Dual Second-order Equivariant Graph Ordinary Differential Equation
Nov 15, 2024



Abstract:Graph Neural Networks (GNNs) with equivariant properties have achieved significant success in modeling complex dynamic systems and molecular properties. However, their expressiveness ability is limited by: (1) Existing methods often overlook the over-smoothing issue caused by traditional GNN models, as well as the gradient explosion or vanishing problems in deep GNNs. (2) Most models operate on first-order information, neglecting that the real world often consists of second-order systems, which further limits the model's representation capabilities. To address these issues, we propose the \textbf{Du}al \textbf{S}econd-order \textbf{E}quivariant \textbf{G}raph \textbf{O}rdinary Differential Equation (\method{}) for equivariant representation. Specifically, \method{} apply the dual second-order equivariant graph ordinary differential equations (Graph ODEs) on graph embeddings and node coordinates, simultaneously. Theoretically, we first prove that \method{} maintains the equivariant property. Furthermore, we provide theoretical insights showing that \method{} effectively alleviates the over-smoothing problem in both feature representation and coordinate update. Additionally, we demonstrate that the proposed \method{} mitigates the exploding and vanishing gradients problem, facilitating the training of deep multi-layer GNNs. Extensive experiments on benchmark datasets validate the superiority of the proposed \method{} compared to baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge