Xuanyu Wang
RPIQ: Residual-Projected Multi-Collaboration Closed-Loop and Single Instance Quantization for Visually Impaired Assistance
Jan 06, 2026Abstract:Visually impaired users face significant challenges in daily information access and real-time environmental perception, and there is an urgent need for intelligent assistive systems with accurate recognition capabilities. Although large-scale models provide effective solutions for perception and reasoning, their practical deployment on assistive devices is severely constrained by excessive memory consumption and high inference costs. Moreover, existing quantization strategies often ignore inter-block error accumulation, leading to degraded model stability. To address these challenges, this study proposes a novel quantization framework -- Residual-Projected Multi-Collaboration Closed-Loop and Single Instance Quantization(RPIQ), whose quantization process adopts a multi-collaborative closed-loop compensation scheme based on Single Instance Calibration and Gauss-Seidel Iterative Quantization. Experiments on various types of large-scale models, including language models such as OPT, Qwen, and LLaMA, as well as vision-language models such as CogVLM2, demonstrate that RPIQ can compress models to 4-bit representation while significantly reducing peak memory consumption (approximately 60%-75% reduction compared to original full-precision models). The method maintains performance highly close to full-precision models across multiple language and visual tasks, and exhibits excellent recognition and reasoning capabilities in key applications such as text understanding and visual question answering in complex scenarios. While verifying the effectiveness of RPIQ for deployment in real assistive systems, this study also advances the computational efficiency and reliability of large models, enabling them to provide visually impaired users with the required information accurately and rapidly.
Deep Pathomic Learning Defines Prognostic Subtypes and Molecular Drivers in Colorectal Cancer
Nov 19, 2025



Abstract:Precise prognostic stratification of colorectal cancer (CRC) remains a major clinical challenge due to its high heterogeneity. The conventional TNM staging system is inadequate for personalized medicine. We aimed to develop and validate a novel multiple instance learning model TDAM-CRC using histopathological whole-slide images for accurate prognostic prediction and to uncover its underlying molecular mechanisms. We trained the model on the TCGA discovery cohort (n=581), validated it in an independent external cohort (n=1031), and further we integrated multi-omics data to improve model interpretability and identify novel prognostic biomarkers. The results demonstrated that the TDAM-CRC achieved robust risk stratification in both cohorts. Its predictive performance significantly outperformed the conventional clinical staging system and multiple state-of-the-art models. The TDAM-CRC risk score was confirmed as an independent prognostic factor in multivariable analysis. Multi-omics analysis revealed that the high-risk subtype is closely associated with metabolic reprogramming and an immunosuppressive tumor microenvironment. Through interaction network analysis, we identified and validated Mitochondrial Ribosomal Protein L37 (MRPL37) as a key hub gene linking deep pathomic features to clinical prognosis. We found that high expression of MRPL37, driven by promoter hypomethylation, serves as an independent biomarker of favorable prognosis. Finally, we constructed a nomogram incorporating the TDAM-CRC risk score and clinical factors to provide a precise and interpretable clinical decision-making tool for CRC patients. Our AI-driven pathological model TDAM-CRC provides a robust tool for improved CRC risk stratification, reveals new molecular targets, and facilitates personalized clinical decision-making.
Scene-Aware Vectorized Memory Multi-Agent Framework with Cross-Modal Differentiated Quantization VLMs for Visually Impaired Assistance
Aug 25, 2025Abstract:This study proposes the dual technological innovation framework, including a cross-modal differ entiated quantization framework for vision-language models (VLMs) and a scene-aware vectorized memory multi-agent system for visually impaired assistance. The modular framework was developed implementing differentiated processing strategies, effectively reducing memory requirements from 38GB to 16GB while maintaining model performance. The multi-agent architecture combines scene classification, vectorized memory, and multimodal interaction, enabling persistent storage and efficient retrieval of scene memories. Through perception-memory-reasoning workflows, the system provides environmental information beyond the current view using historical memories. Experiments show the quantized 19B-parameter model only experiences a 2.05% performance drop on MMBench and maintains 63.7 accuracy on OCR-VQA (original: 64.9), outperforming smaller models with equivalent memory requirements like the Molmo-7B series. The system maintains response latency between 2.83-3.52 seconds from scene analysis to initial speech output, substantially faster than non-streaming methods. This research advances computational efficiency and assistive technology, offering visually impaired users comprehensive real-time assistance in scene perception, text recognition, and navigation.
Subspecialty-Specific Foundation Model for Intelligent Gastrointestinal Pathology
May 28, 2025Abstract:Gastrointestinal (GI) diseases represent a clinically significant burden, necessitating precise diagnostic approaches to optimize patient outcomes. Conventional histopathological diagnosis, heavily reliant on the subjective interpretation of pathologists, suffers from limited reproducibility and diagnostic variability. To overcome these limitations and address the lack of pathology-specific foundation models for GI diseases, we develop Digepath, a specialized foundation model for GI pathology. Our framework introduces a dual-phase iterative optimization strategy combining pretraining with fine-screening, specifically designed to address the detection of sparsely distributed lesion areas in whole-slide images. Digepath is pretrained on more than 353 million image patches from over 200,000 hematoxylin and eosin-stained slides of GI diseases. It attains state-of-the-art performance on 33 out of 34 tasks related to GI pathology, including pathological diagnosis, molecular prediction, gene mutation prediction, and prognosis evaluation, particularly in diagnostically ambiguous cases and resolution-agnostic tissue classification.We further translate the intelligent screening module for early GI cancer and achieve near-perfect 99.6% sensitivity across 9 independent medical institutions nationwide. The outstanding performance of Digepath highlights its potential to bridge critical gaps in histopathological practice. This work not only advances AI-driven precision pathology for GI diseases but also establishes a transferable paradigm for other pathology subspecialties.
Demystifying Workload Imbalances in Large Transformer Model Training over Variable-length Sequences
Dec 10, 2024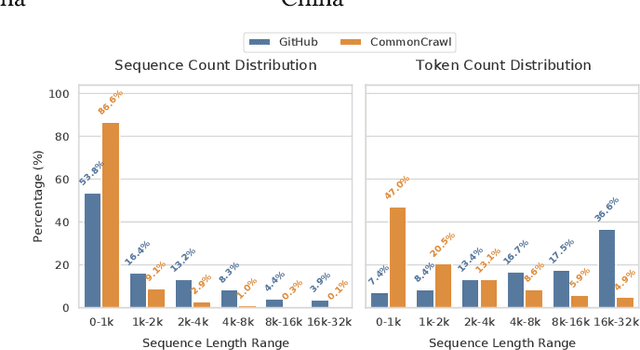
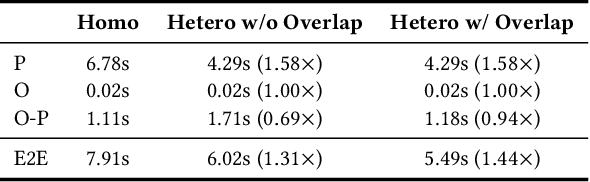
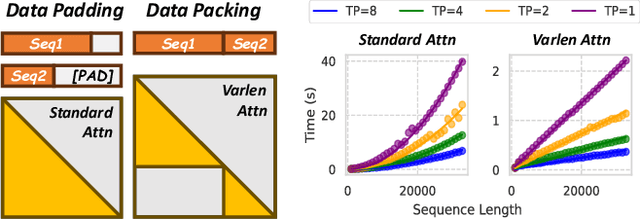
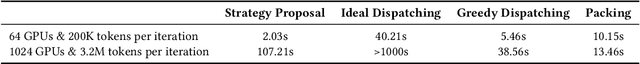
Abstract:To optimize large Transformer model training, efficient parallel computing and advanced data management are essential. However, current methods often assume a stable and uniform training workload, neglecting imbalances in data sampling and packing that can impede performance. Specifically, data sampling imbalance arises from uneven sequence length distribution of the training data, while data packing imbalance stems from the discrepancy between the linear memory complexity and quadratic time complexity of the attention mechanism. To address these imbalance issues, we develop Hydraulis, which jointly optimizes the parallel strategies and data assignment. For one thing, we introduce large model training with dynamic heterogeneous parallel strategies in response to the sequence length variations within and across training iterations. For another, we devise a two-stage data assignment approach, which strikes a good balance in terms of the training workloads both within and across model replicas. Empirical results demonstrate that Hydraulis outperforms existing systems by 1.32-2.66 times.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge