Efficient Approximations of Complete Interatomic Potentials for Crystal Property Prediction
Jun 21, 2023Yuchao Lin, Keqiang Yan, Youzhi Luo, Yi Liu, Xiaoning Qian, Shuiwang Ji
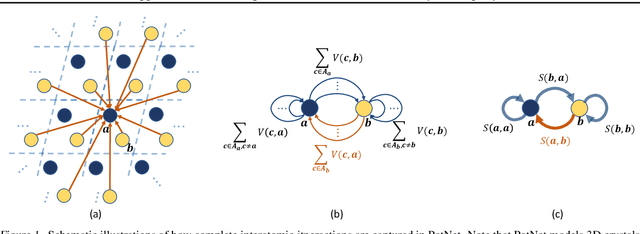
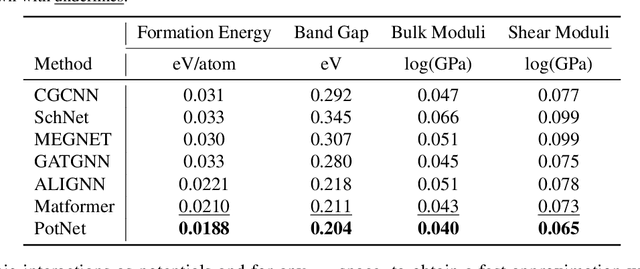
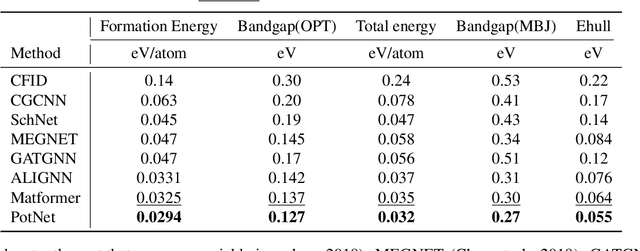
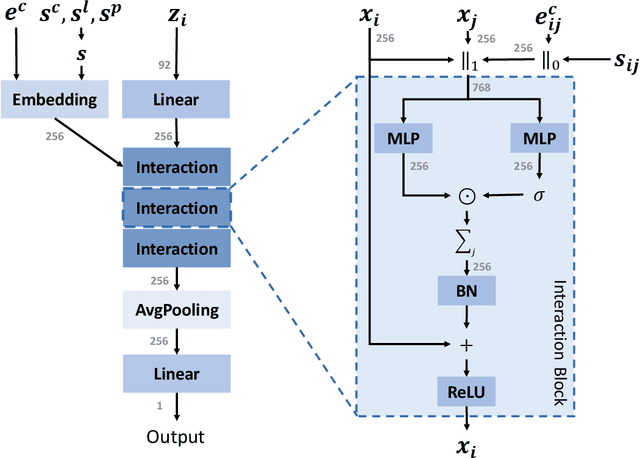
We study property prediction for crystal materials. A crystal structure consists of a minimal unit cell that is repeated infinitely in 3D space. How to accurately represent such repetitive structures in machine learning models remains unresolved. Current methods construct graphs by establishing edges only between nearby nodes, thereby failing to faithfully capture infinite repeating patterns and distant interatomic interactions. In this work, we propose several innovations to overcome these limitations. First, we propose to model physics-principled interatomic potentials directly instead of only using distances as in many existing methods. These potentials include the Coulomb potential, London dispersion potential, and Pauli repulsion potential. Second, we model the complete set of potentials among all atoms, instead of only between nearby atoms as in existing methods. This is enabled by our approximations of infinite potential summations with provable error bounds. We further develop efficient algorithms to compute the approximations. Finally, we propose to incorporate our computations of complete interatomic potentials into message passing neural networks for representation learning. We perform experiments on the JARVIS and Materials Project benchmarks for evaluation. Results show that the use of interatomic potentials and complete interatomic potentials leads to consistent performance improvements with reasonable computational costs. Our code is publicly available as part of the AIRS library (https://github.com/divelab/AIRS).
QH9: A Quantum Hamiltonian Prediction Benchmark for QM9 Molecules
Jun 15, 2023Haiyang Yu, Meng Liu, Youzhi Luo, Alex Strasser, Xiaofeng Qian, Xiaoning Qian, Shuiwang Ji
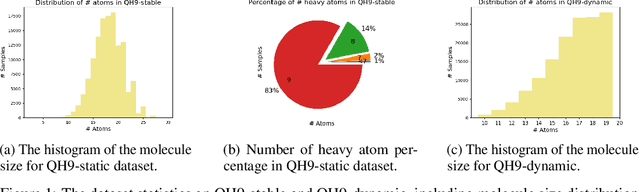
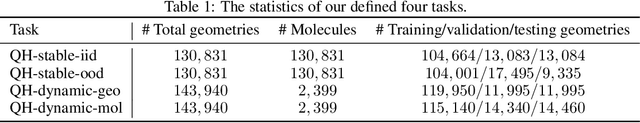
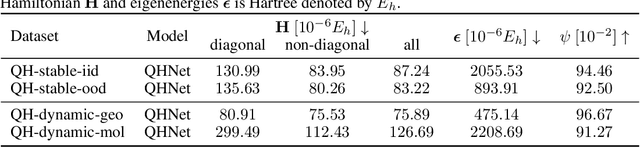

Supervised machine learning approaches have been increasingly used in accelerating electronic structure prediction as surrogates of first-principle computational methods, such as density functional theory (DFT). While numerous quantum chemistry datasets focus on chemical properties and atomic forces, the ability to achieve accurate and efficient prediction of the Hamiltonian matrix is highly desired, as it is the most important and fundamental physical quantity that determines the quantum states of physical systems and chemical properties. In this work, we generate a new Quantum Hamiltonian dataset, named as QH9, to provide precise Hamiltonian matrices for 2,399 molecular dynamics trajectories and 130,831 stable molecular geometries, based on the QM9 dataset. By designing benchmark tasks with various molecules, we show that current machine learning models have the capacity to predict Hamiltonian matrices for arbitrary molecules. Both the QH9 dataset and the baseline models are provided to the community through an open-source benchmark, which can be highly valuable for developing machine learning methods and accelerating molecular and materials design for scientific and technological applications. Our benchmark is publicly available at https://github.com/divelab/AIRS/tree/main/OpenDFT/QHBench.
Efficient and Equivariant Graph Networks for Predicting Quantum Hamiltonian
Jun 08, 2023Haiyang Yu, Zhao Xu, Xiaofeng Qian, Xiaoning Qian, Shuiwang Ji
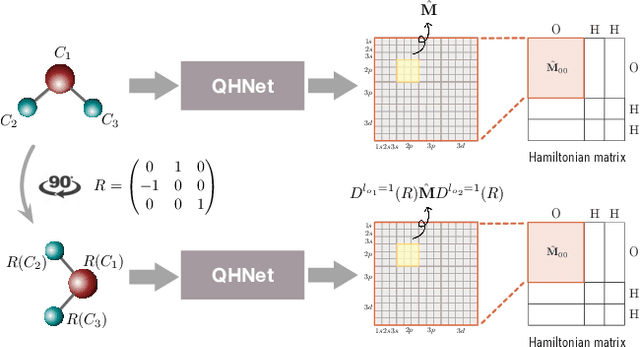
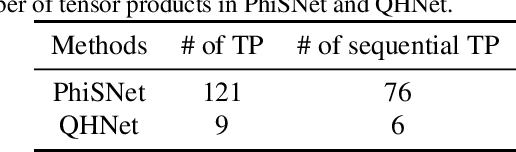
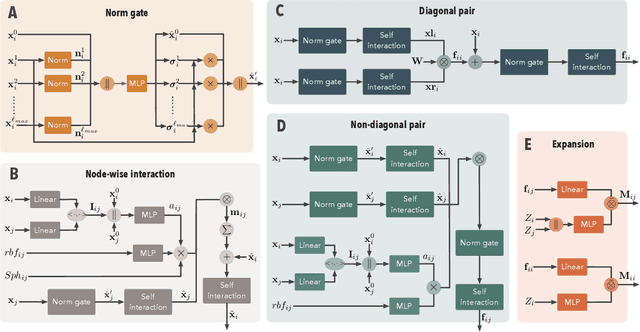
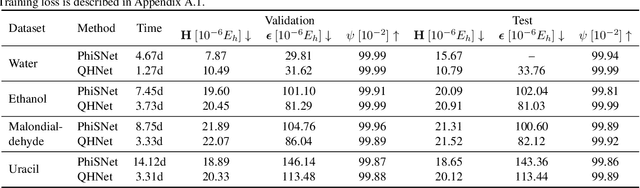
We consider the prediction of the Hamiltonian matrix, which finds use in quantum chemistry and condensed matter physics. Efficiency and equivariance are two important, but conflicting factors. In this work, we propose a SE(3)-equivariant network, named QHNet, that achieves efficiency and equivariance. Our key advance lies at the innovative design of QHNet architecture, which not only obeys the underlying symmetries, but also enables the reduction of number of tensor products by 92\%. In addition, QHNet prevents the exponential growth of channel dimension when more atom types are involved. We perform experiments on MD17 datasets, including four molecular systems. Experimental results show that our QHNet can achieve comparable performance to the state of the art methods at a significantly faster speed. Besides, our QHNet consumes 50\% less memory due to its streamlined architecture. Our code is publicly available as part of the AIRS library (\url{https://github.com/divelab/AIRS}).
A Latent Diffusion Model for Protein Structure Generation
May 06, 2023Cong Fu, Keqiang Yan, Limei Wang, Wing Yee Au, Michael McThrow, Tao Komikado, Koji Maruhashi, Kanji Uchino, Xiaoning Qian, Shuiwang Ji
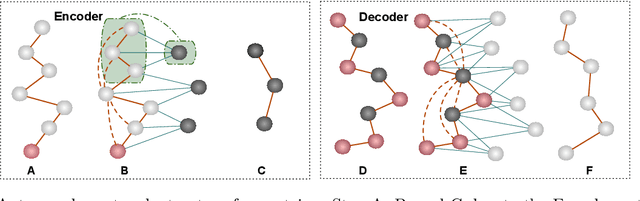
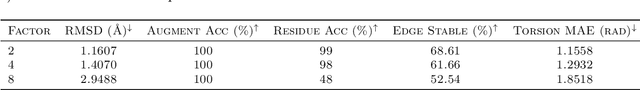
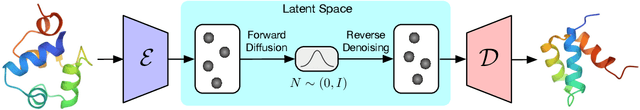
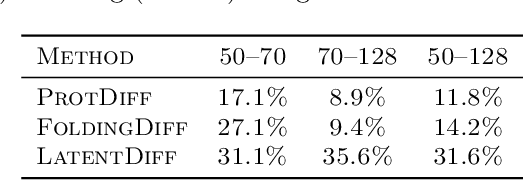
Proteins are complex biomolecules that perform a variety of crucial functions within living organisms. Designing and generating novel proteins can pave the way for many future synthetic biology applications, including drug discovery. However, it remains a challenging computational task due to the large modeling space of protein structures. In this study, we propose a latent diffusion model that can reduce the complexity of protein modeling while flexibly capturing the distribution of natural protein structures in a condensed latent space. Specifically, we propose an equivariant protein autoencoder that embeds proteins into a latent space and then uses an equivariant diffusion model to learn the distribution of the latent protein representations. Experimental results demonstrate that our method can effectively generate novel protein backbone structures with high designability and efficiency.
Comprehensive analysis of gene expression profiles to radiation exposure reveals molecular signatures of low-dose radiation response
Jan 03, 2023Xihaier Luo, Sean McCorkle, Gilchan Park, Vanessa Lopez-Marrero, Shinjae Yoo, Edward R. Dougherty, Xiaoning Qian, Francis J. Alexander, Byung-Jun Yoon
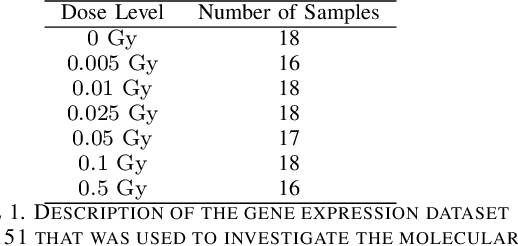
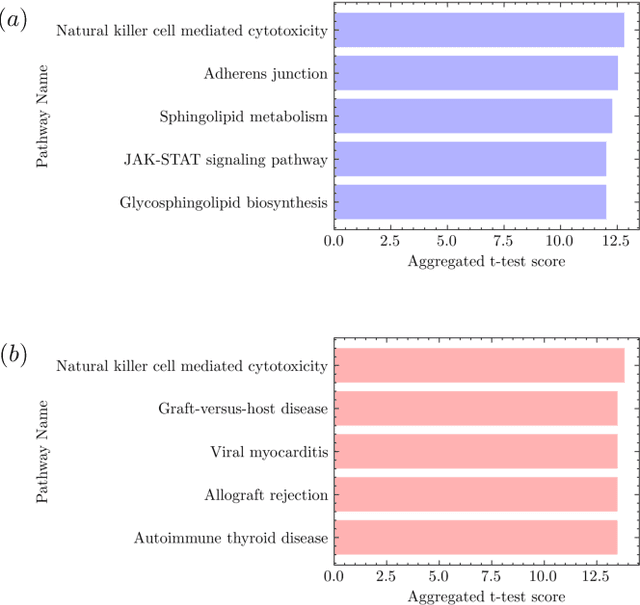
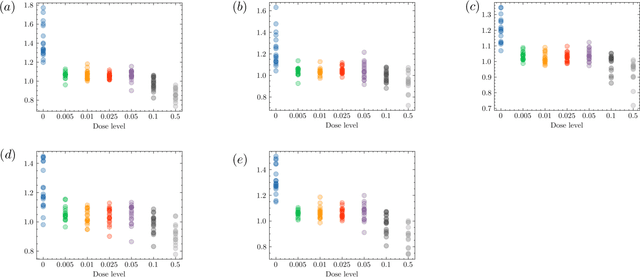
There are various sources of ionizing radiation exposure, where medical exposure for radiation therapy or diagnosis is the most common human-made source. Understanding how gene expression is modulated after ionizing radiation exposure and investigating the presence of any dose-dependent gene expression patterns have broad implications for health risks from radiotherapy, medical radiation diagnostic procedures, as well as other environmental exposure. In this paper, we perform a comprehensive pathway-based analysis of gene expression profiles in response to low-dose radiation exposure, in order to examine the potential mechanism of gene regulation underlying such responses. To accomplish this goal, we employ a statistical framework to determine whether a specific group of genes belonging to a known pathway display coordinated expression patterns that are modulated in a manner consistent with the radiation level. Findings in our study suggest that there exist complex yet consistent signatures that reflect the molecular response to radiation exposure, which differ between low-dose and high-dose radiation.
DynImp: Dynamic Imputation for Wearable Sensing Data Through Sensory and Temporal Relatedness
Sep 26, 2022Zepeng Huo, Taowei Ji, Yifei Liang, Shuai Huang, Zhangyang Wang, Xiaoning Qian, Bobak Mortazavi
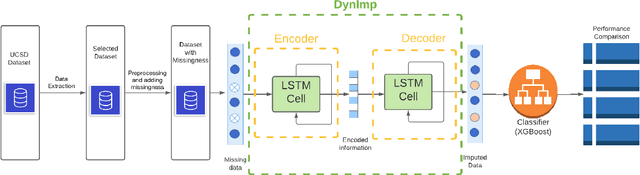
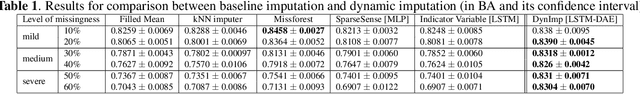
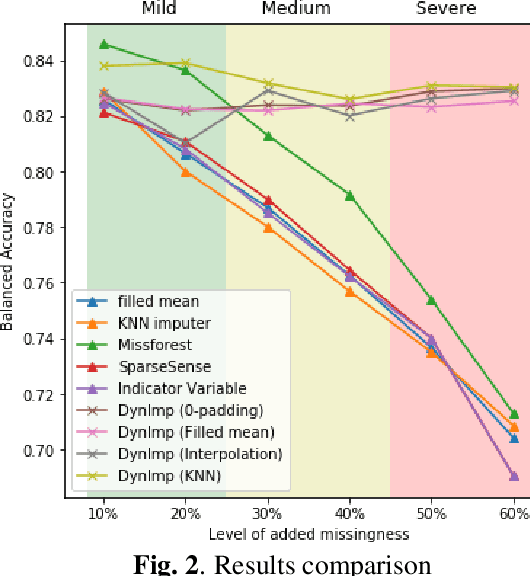
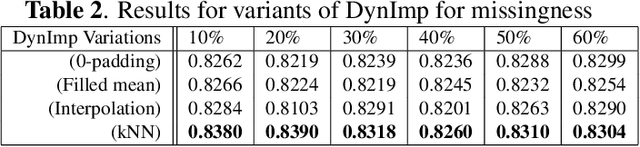
In wearable sensing applications, data is inevitable to be irregularly sampled or partially missing, which pose challenges for any downstream application. An unique aspect of wearable data is that it is time-series data and each channel can be correlated to another one, such as x, y, z axis of accelerometer. We argue that traditional methods have rarely made use of both times-series dynamics of the data as well as the relatedness of the features from different sensors. We propose a model, termed as DynImp, to handle different time point's missingness with nearest neighbors along feature axis and then feeding the data into a LSTM-based denoising autoencoder which can reconstruct missingness along the time axis. We experiment the model on the extreme missingness scenario ($>50\%$ missing rate) which has not been widely tested in wearable data. Our experiments on activity recognition show that the method can exploit the multi-modality features from related sensors and also learn from history time-series dynamics to reconstruct the data under extreme missingness.
Density-Aware Personalized Training for Risk Prediction in Imbalanced Medical Data
Jul 29, 2022Zepeng Huo, Xiaoning Qian, Shuai Huang, Zhangyang Wang, Bobak J. Mortazavi
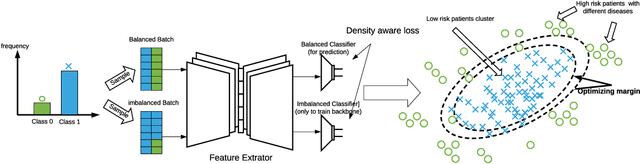

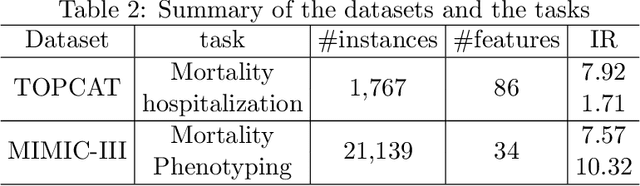
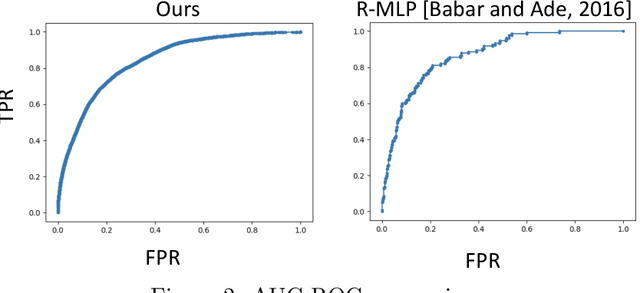
Medical events of interest, such as mortality, often happen at a low rate in electronic medical records, as most admitted patients survive. Training models with this imbalance rate (class density discrepancy) may lead to suboptimal prediction. Traditionally this problem is addressed through ad-hoc methods such as resampling or reweighting but performance in many cases is still limited. We propose a framework for training models for this imbalance issue: 1) we first decouple the feature extraction and classification process, adjusting training batches separately for each component to mitigate bias caused by class density discrepancy; 2) we train the network with both a density-aware loss and a learnable cost matrix for misclassifications. We demonstrate our model's improved performance in real-world medical datasets (TOPCAT and MIMIC-III) to show improved AUC-ROC, AUC-PRC, Brier Skill Score compared with the baselines in the domain.
VFDS: Variational Foresight Dynamic Selection in Bayesian Neural Networks for Efficient Human Activity Recognition
Mar 31, 2022Randy Ardywibowo, Shahin Boluki, Zhangyang Wang, Bobak Mortazavi, Shuai Huang, Xiaoning Qian
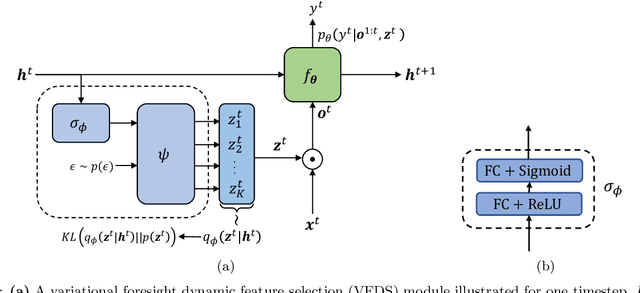
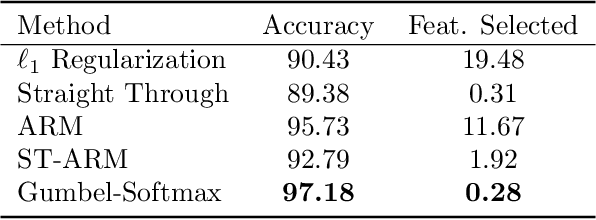
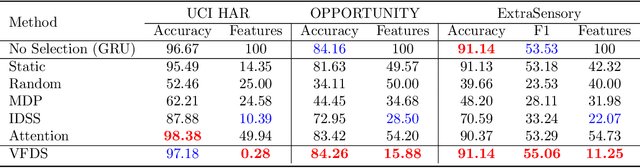
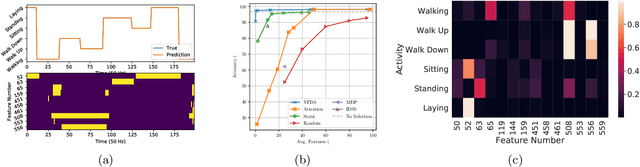
In many machine learning tasks, input features with varying degrees of predictive capability are acquired at varying costs. In order to optimize the performance-cost trade-off, one would select features to observe a priori. However, given the changing context with previous observations, the subset of predictive features to select may change dynamically. Therefore, we face the challenging new problem of foresight dynamic selection (FDS): finding a dynamic and light-weight policy to decide which features to observe next, before actually observing them, for overall performance-cost trade-offs. To tackle FDS, this paper proposes a Bayesian learning framework of Variational Foresight Dynamic Selection (VFDS). VFDS learns a policy that selects the next feature subset to observe, by optimizing a variational Bayesian objective that characterizes the trade-off between model performance and feature cost. At its core is an implicit variational distribution on binary gates that are dependent on previous observations, which will select the next subset of features to observe. We apply VFDS on the Human Activity Recognition (HAR) task where the performance-cost trade-off is critical in its practice. Extensive results demonstrate that VFDS selects different features under changing contexts, notably saving sensory costs while maintaining or improving the HAR accuracy. Moreover, the features that VFDS dynamically select are shown to be interpretable and associated with the different activity types. We will release the code.
MoReL: Multi-omics Relational Learning
Mar 15, 2022Arman Hasanzadeh, Ehsan Hajiramezanali, Nick Duffield, Xiaoning Qian
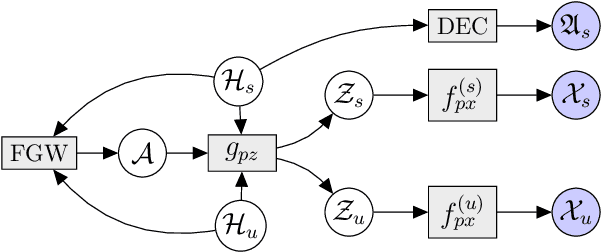
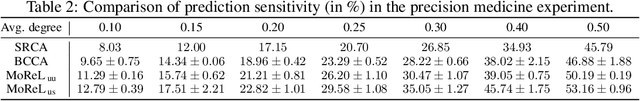
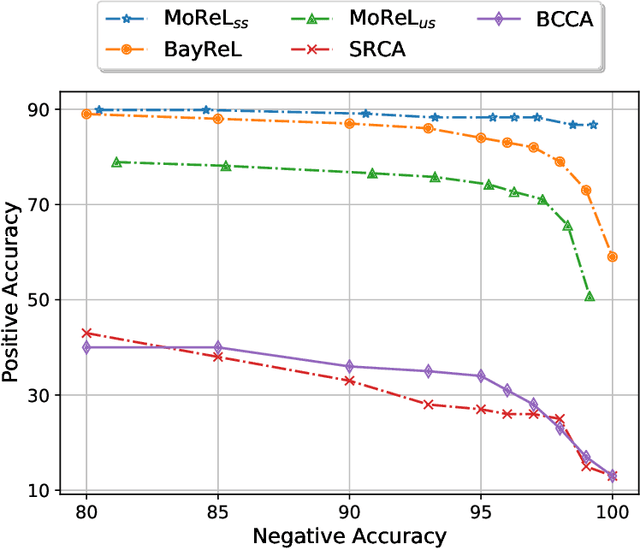
Multi-omics data analysis has the potential to discover hidden molecular interactions, revealing potential regulatory and/or signal transduction pathways for cellular processes of interest when studying life and disease systems. One of critical challenges when dealing with real-world multi-omics data is that they may manifest heterogeneous structures and data quality as often existing data may be collected from different subjects under different conditions for each type of omics data. We propose a novel deep Bayesian generative model to efficiently infer a multi-partite graph encoding molecular interactions across such heterogeneous views, using a fused Gromov-Wasserstein (FGW) regularization between latent representations of corresponding views for integrative analysis. With such an optimal transport regularization in the deep Bayesian generative model, it not only allows incorporating view-specific side information, either with graph-structured or unstructured data in different views, but also increases the model flexibility with the distribution-based regularization. This allows efficient alignment of heterogeneous latent variable distributions to derive reliable interaction predictions compared to the existing point-based graph embedding methods. Our experiments on several real-world datasets demonstrate enhanced performance of MoReL in inferring meaningful interactions compared to existing baselines.
COVID-Datathon: Biomarker identification for COVID-19 severity based on BALF scRNA-seq data
Oct 11, 2021Seyednami Niyakan, Xiaoning Qian
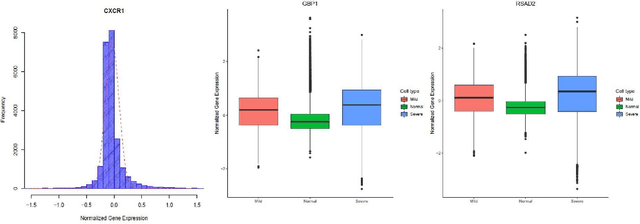
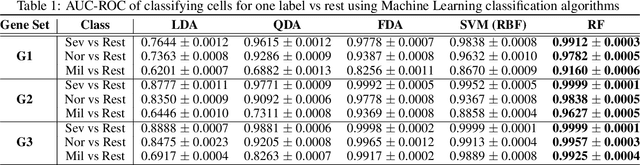
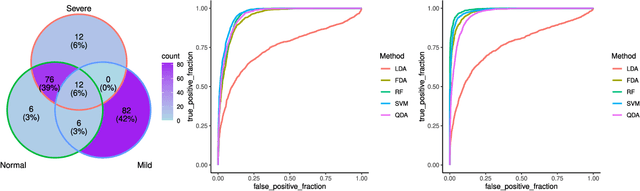
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emergence began in late 2019 and has since spread rapidly worldwide. The characteristics of respiratory immune response to this emerging virus is not clear. Recently, Single-cell RNA sequencing (scRNA-seq) transcriptome profiling of Bronchoalveolar lavage fluid (BALF) cells has been done to elucidate the potential mechanisms underlying in COVID-19. With the aim of better utilizing this atlas of BALF cells in response to the virus, here we propose a bioinformatics pipeline to identify candidate biomarkers of COVID-19 severity, which may help characterize BALF cells to have better mechanistic understanding of SARS-CoV-2 infection. The proposed pipeline is implemented in R and is available at https://github.com/namini94/scBALF_Hackathon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge