Xiaofeng Liu
MRecGen: Multimodal Appropriate Reaction Generator
Jul 05, 2023Abstract:Verbal and non-verbal human reaction generation is a challenging task, as different reactions could be appropriate for responding to the same behaviour. This paper proposes the first multiple and multimodal (verbal and nonverbal) appropriate human reaction generation framework that can generate appropriate and realistic human-style reactions (displayed in the form of synchronised text, audio and video streams) in response to an input user behaviour. This novel technique can be applied to various human-computer interaction scenarios by generating appropriate virtual agent/robot behaviours. Our demo is available at \url{https://github.com/SSYSteve/MRecGen}.
Incremental Learning for Heterogeneous Structure Segmentation in Brain Tumor MRI
May 30, 2023



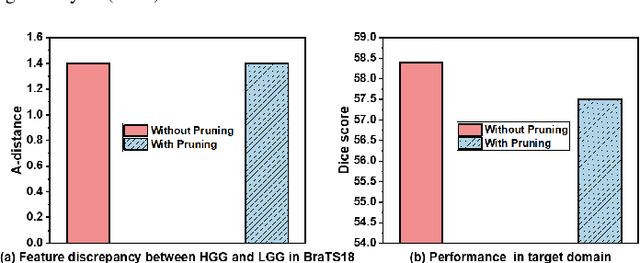
Abstract:Deep learning (DL) models for segmenting various anatomical structures have achieved great success via a static DL model that is trained in a single source domain. Yet, the static DL model is likely to perform poorly in a continually evolving environment, requiring appropriate model updates. In an incremental learning setting, we would expect that well-trained static models are updated, following continually evolving target domain data -- e.g., additional lesions or structures of interest -- collected from different sites, without catastrophic forgetting. This, however, poses challenges, due to distribution shifts, additional structures not seen during the initial model training, and the absence of training data in a source domain. To address these challenges, in this work, we seek to progressively evolve an ``off-the-shelf" trained segmentation model to diverse datasets with additional anatomical categories in a unified manner. Specifically, we first propose a divergence-aware dual-flow module with balanced rigidity and plasticity branches to decouple old and new tasks, which is guided by continuous batch renormalization. Then, a complementary pseudo-label training scheme with self-entropy regularized momentum MixUp decay is developed for adaptive network optimization. We evaluated our framework on a brain tumor segmentation task with continually changing target domains -- i.e., new MRI scanners/modalities with incremental structures. Our framework was able to well retain the discriminability of previously learned structures, hence enabling the realistic life-long segmentation model extension along with the widespread accumulation of big medical data.
Attentive Continuous Generative Self-training for Unsupervised Domain Adaptive Medical Image Translation
May 23, 2023Abstract:Self-training is an important class of unsupervised domain adaptation (UDA) approaches that are used to mitigate the problem of domain shift, when applying knowledge learned from a labeled source domain to unlabeled and heterogeneous target domains. While self-training-based UDA has shown considerable promise on discriminative tasks, including classification and segmentation, through reliable pseudo-label filtering based on the maximum softmax probability, there is a paucity of prior work on self-training-based UDA for generative tasks, including image modality translation. To fill this gap, in this work, we seek to develop a generative self-training (GST) framework for domain adaptive image translation with continuous value prediction and regression objectives. Specifically, we quantify both aleatoric and epistemic uncertainties within our GST using variational Bayes learning to measure the reliability of synthesized data. We also introduce a self-attention scheme that de-emphasizes the background region to prevent it from dominating the training process. The adaptation is then carried out by an alternating optimization scheme with target domain supervision that focuses attention on the regions with reliable pseudo-labels. We evaluated our framework on two cross-scanner/center, inter-subject translation tasks, including tagged-to-cine magnetic resonance (MR) image translation and T1-weighted MR-to-fractional anisotropy translation. Extensive validations with unpaired target domain data showed that our GST yielded superior synthesis performance in comparison to adversarial training UDA methods.
NFI$_2$: Learning Noise-Free Illuminance-Interpolator for Unsupervised Low-Light Image Enhancement
May 17, 2023Abstract:Low-light situations severely restrict the pursuit of aesthetic quality in consumer photography. Although many efforts are devoted to designing heuristics, it is generally mired in a shallow spiral of tedium, such as piling up complex network architectures and empirical strategies. How to delve into the essential physical principles of illumination compensation has been neglected. Following the way of simplifying the complexity, this paper innovatively proposes a simple and efficient Noise-Free Illumination Interpolator (NFI$_2$). According to the constraint principle of illuminance and reflectance within a limited dynamic range, as a prior knowledge in the recovery process, we construct a learnable illuminance interpolator and thereby compensating for non-uniform lighting. With the intention of adapting denoising without annotated data, we design a self-calibrated denoiser with the intrinsic image properties to acquire noise-free low-light images. Starting from the properties of natural image manifolds, a self-regularized recovery loss is introduced as a way to encourage more natural and realistic reflectance map. The model architecture and training losses, guided by prior knowledge, complement and benefit each other, forming a powerful unsupervised leaning framework. Comprehensive experiments demonstrate that the proposed algorithm produces competitive qualitative and quantitative results while maintaining favorable generalization capability in unknown real-world scenarios.
Posterior Estimation Using Deep Learning: A Simulation Study of Compartmental Modeling in Dynamic PET
Mar 17, 2023Abstract:Background: In medical imaging, images are usually treated as deterministic, while their uncertainties are largely underexplored. Purpose: This work aims at using deep learning to efficiently estimate posterior distributions of imaging parameters, which in turn can be used to derive the most probable parameters as well as their uncertainties. Methods: Our deep learning-based approaches are based on a variational Bayesian inference framework, which is implemented using two different deep neural networks based on conditional variational auto-encoder (CVAE), CVAE-dual-encoder and CVAE-dual-decoder. The conventional CVAE framework, i.e., CVAE-vanilla, can be regarded as a simplified case of these two neural networks. We applied these approaches to a simulation study of dynamic brain PET imaging using a reference region-based kinetic model. Results: In the simulation study, we estimated posterior distributions of PET kinetic parameters given a measurement of time-activity curve. Our proposed CVAE-dual-encoder and CVAE-dual-decoder yield results that are in good agreement with the asymptotically unbiased posterior distributions sampled by Markov Chain Monte Carlo (MCMC). The CVAE-vanilla can also be used for estimating posterior distributions, although it has an inferior performance to both CVAE-dual-encoder and CVAE-dual-decoder. Conclusions: We have evaluated the performance of our deep learning approaches for estimating posterior distributions in dynamic brain PET. Our deep learning approaches yield posterior distributions, which are in good agreement with unbiased distributions estimated by MCMC. All these neural networks have different characteristics and can be chosen by the user for specific applications. The proposed methods are general and can be adapted to other problems.
Synthesizing audio from tongue motion during speech using tagged MRI via transformer
Feb 14, 2023Abstract:Investigating the relationship between internal tissue point motion of the tongue and oropharyngeal muscle deformation measured from tagged MRI and intelligible speech can aid in advancing speech motor control theories and developing novel treatment methods for speech related-disorders. However, elucidating the relationship between these two sources of information is challenging, due in part to the disparity in data structure between spatiotemporal motion fields (i.e., 4D motion fields) and one-dimensional audio waveforms. In this work, we present an efficient encoder-decoder translation network for exploring the predictive information inherent in 4D motion fields via 2D spectrograms as a surrogate of the audio data. Specifically, our encoder is based on 3D convolutional spatial modeling and transformer-based temporal modeling. The extracted features are processed by an asymmetric 2D convolution decoder to generate spectrograms that correspond to 4D motion fields. Furthermore, we incorporate a generative adversarial training approach into our framework to further improve synthesis quality on our generated spectrograms. We experiment on 63 paired motion field sequences and speech waveforms, demonstrating that our framework enables the generation of clear audio waveforms from a sequence of motion fields. Thus, our framework has the potential to improve our understanding of the relationship between these two modalities and inform the development of treatments for speech disorders.
Successive Subspace Learning for Cardiac Disease Classification with Two-phase Deformation Fields from Cine MRI
Jan 21, 2023Abstract:Cardiac cine magnetic resonance imaging (MRI) has been used to characterize cardiovascular diseases (CVD), often providing a noninvasive phenotyping tool.~While recently flourished deep learning based approaches using cine MRI yield accurate characterization results, the performance is often degraded by small training samples. In addition, many deep learning models are deemed a ``black box," for which models remain largely elusive in how models yield a prediction and how reliable they are. To alleviate this, this work proposes a lightweight successive subspace learning (SSL) framework for CVD classification, based on an interpretable feedforward design, in conjunction with a cardiac atlas. Specifically, our hierarchical SSL model is based on (i) neighborhood voxel expansion, (ii) unsupervised subspace approximation, (iii) supervised regression, and (iv) multi-level feature integration. In addition, using two-phase 3D deformation fields, including end-diastolic and end-systolic phases, derived between the atlas and individual subjects as input offers objective means of assessing CVD, even with small training samples. We evaluate our framework on the ACDC2017 database, comprising one healthy group and four disease groups. Compared with 3D CNN-based approaches, our framework achieves superior classification performance with 140$\times$ fewer parameters, which supports its potential value in clinical use.
Differentiable modeling to unify machine learning and physical models and advance Geosciences
Jan 10, 2023



Abstract:Process-Based Modeling (PBM) and Machine Learning (ML) are often perceived as distinct paradigms in the geosciences. Here we present differentiable geoscientific modeling as a powerful pathway toward dissolving the perceived barrier between them and ushering in a paradigm shift. For decades, PBM offered benefits in interpretability and physical consistency but struggled to efficiently leverage large datasets. ML methods, especially deep networks, presented strong predictive skills yet lacked the ability to answer specific scientific questions. While various methods have been proposed for ML-physics integration, an important underlying theme -- differentiable modeling -- is not sufficiently recognized. Here we outline the concepts, applicability, and significance of differentiable geoscientific modeling (DG). "Differentiable" refers to accurately and efficiently calculating gradients with respect to model variables, critically enabling the learning of high-dimensional unknown relationships. DG refers to a range of methods connecting varying amounts of prior knowledge to neural networks and training them together, capturing a different scope than physics-guided machine learning and emphasizing first principles. Preliminary evidence suggests DG offers better interpretability and causality than ML, improved generalizability and extrapolation capability, and strong potential for knowledge discovery, while approaching the performance of purely data-driven ML. DG models require less training data while scaling favorably in performance and efficiency with increasing amounts of data. With DG, geoscientists may be better able to frame and investigate questions, test hypotheses, and discover unrecognized linkages.
Infrared Image Super-Resolution: Systematic Review, and Future Trends
Dec 22, 2022Abstract:Image Super-Resolution (SR) is essential for a wide range of computer vision and image processing tasks. Investigating infrared (IR) image (or thermal images) super-resolution is a continuing concern within the development of deep learning. This survey aims to provide a comprehensive perspective of IR image super-resolution, including its applications, hardware imaging system dilemmas, and taxonomy of image processing methodologies. In addition, the datasets and evaluation metrics in IR image super-resolution tasks are also discussed. Furthermore, the deficiencies in current technologies and possible promising directions for the community to explore are highlighted. To cope with the rapid development in this field, we intend to regularly update the relevant excellent work at \url{https://github.com/yongsongH/Infrared_Image_SR_Survey
Memory Consistent Unsupervised Off-the-Shelf Model Adaptation for Source-Relaxed Medical Image Segmentation
Sep 16, 2022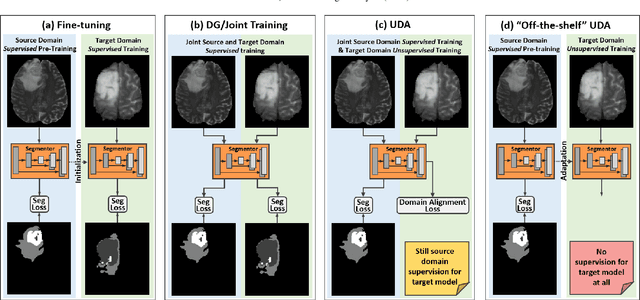
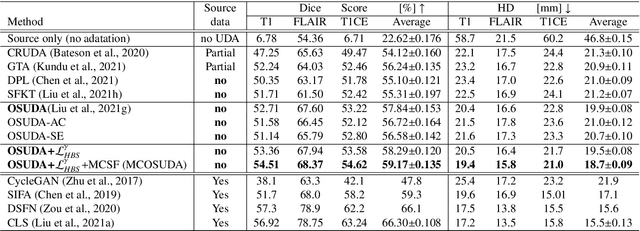
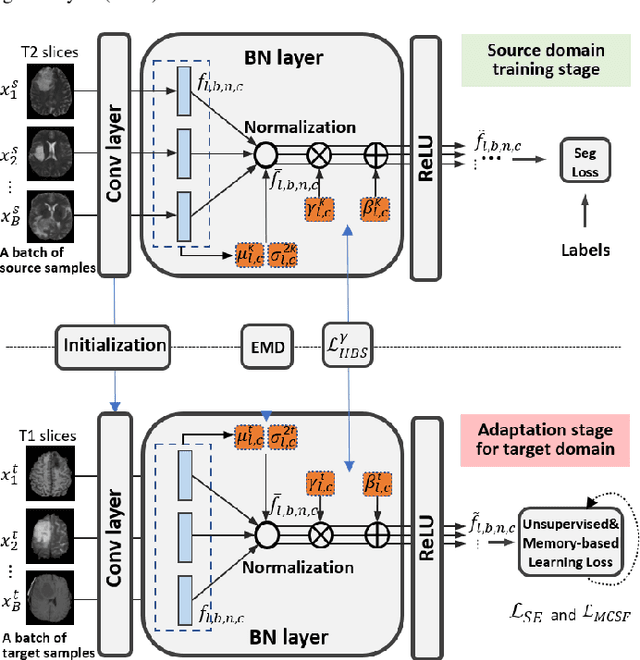
Abstract:Unsupervised domain adaptation (UDA) has been a vital protocol for migrating information learned from a labeled source domain to facilitate the implementation in an unlabeled heterogeneous target domain. Although UDA is typically jointly trained on data from both domains, accessing the labeled source domain data is often restricted, due to concerns over patient data privacy or intellectual property. To sidestep this, we propose "off-the-shelf (OS)" UDA (OSUDA), aimed at image segmentation, by adapting an OS segmentor trained in a source domain to a target domain, in the absence of source domain data in adaptation. Toward this goal, we aim to develop a novel batch-wise normalization (BN) statistics adaptation framework. In particular, we gradually adapt the domain-specific low-order BN statistics, e.g., mean and variance, through an exponential momentum decay strategy, while explicitly enforcing the consistency of the domain shareable high-order BN statistics, e.g., scaling and shifting factors, via our optimization objective. We also adaptively quantify the channel-wise transferability to gauge the importance of each channel, via both low-order statistics divergence and a scaling factor.~Furthermore, we incorporate unsupervised self-entropy minimization into our framework to boost performance alongside a novel queued, memory-consistent self-training strategy to utilize the reliable pseudo label for stable and efficient unsupervised adaptation. We evaluated our OSUDA-based framework on both cross-modality and cross-subtype brain tumor segmentation and cardiac MR to CT segmentation tasks. Our experimental results showed that our memory consistent OSUDA performs better than existing source-relaxed UDA methods and yields similar performance to UDA methods with source data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge