Tommaso Biancalani
Joint Embedding vs Reconstruction: Provable Benefits of Latent Space Prediction for Self Supervised Learning
May 18, 2025



Abstract:Reconstruction and joint embedding have emerged as two leading paradigms in Self Supervised Learning (SSL). Reconstruction methods focus on recovering the original sample from a different view in input space. On the other hand, joint embedding methods align the representations of different views in latent space. Both approaches offer compelling advantages, yet practitioners lack clear guidelines for choosing between them. In this work, we unveil the core mechanisms that distinguish each paradigm. By leveraging closed form solutions for both approaches, we precisely characterize how the view generation process, e.g. data augmentation, impacts the learned representations. We then demonstrate that, unlike supervised learning, both SSL paradigms require a minimal alignment between augmentations and irrelevant features to achieve asymptotic optimality with increasing sample size. Our findings indicate that in scenarios where these irrelevant features have a large magnitude, joint embedding methods are preferable because they impose a strictly weaker alignment condition compared to reconstruction based methods. These results not only clarify the trade offs between the two paradigms but also substantiate the empirical success of joint embedding approaches on real world challenging datasets.
Dynamic Search for Inference-Time Alignment in Diffusion Models
Mar 03, 2025



Abstract:Diffusion models have shown promising generative capabilities across diverse domains, yet aligning their outputs with desired reward functions remains a challenge, particularly in cases where reward functions are non-differentiable. Some gradient-free guidance methods have been developed, but they often struggle to achieve optimal inference-time alignment. In this work, we newly frame inference-time alignment in diffusion as a search problem and propose Dynamic Search for Diffusion (DSearch), which subsamples from denoising processes and approximates intermediate node rewards. It also dynamically adjusts beam width and tree expansion to efficiently explore high-reward generations. To refine intermediate decisions, DSearch incorporates adaptive scheduling based on noise levels and a lookahead heuristic function. We validate DSearch across multiple domains, including biological sequence design, molecular optimization, and image generation, demonstrating superior reward optimization compared to existing approaches.
Contextualizing biological perturbation experiments through language
Feb 28, 2025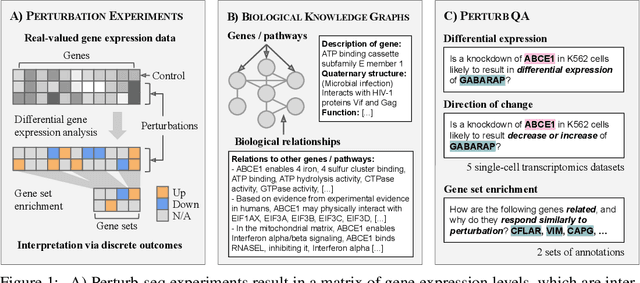
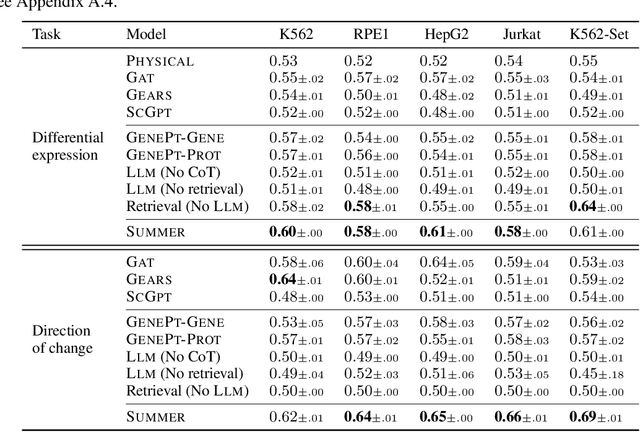
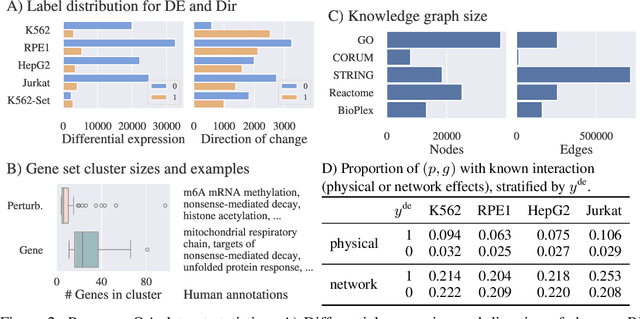
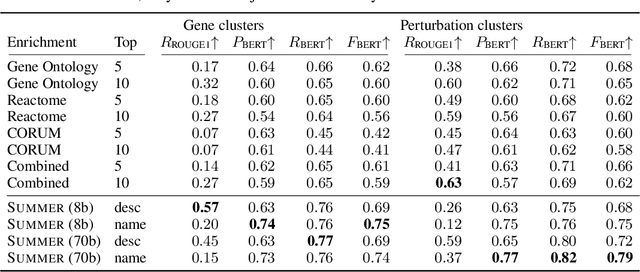
Abstract:High-content perturbation experiments allow scientists to probe biomolecular systems at unprecedented resolution, but experimental and analysis costs pose significant barriers to widespread adoption. Machine learning has the potential to guide efficient exploration of the perturbation space and extract novel insights from these data. However, current approaches neglect the semantic richness of the relevant biology, and their objectives are misaligned with downstream biological analyses. In this paper, we hypothesize that large language models (LLMs) present a natural medium for representing complex biological relationships and rationalizing experimental outcomes. We propose PerturbQA, a benchmark for structured reasoning over perturbation experiments. Unlike current benchmarks that primarily interrogate existing knowledge, PerturbQA is inspired by open problems in perturbation modeling: prediction of differential expression and change of direction for unseen perturbations, and gene set enrichment. We evaluate state-of-the-art machine learning and statistical approaches for modeling perturbations, as well as standard LLM reasoning strategies, and we find that current methods perform poorly on PerturbQA. As a proof of feasibility, we introduce Summer (SUMMarize, retrievE, and answeR, a simple, domain-informed LLM framework that matches or exceeds the current state-of-the-art. Our code and data are publicly available at https://github.com/genentech/PerturbQA.
Reward-Guided Iterative Refinement in Diffusion Models at Test-Time with Applications to Protein and DNA Design
Feb 20, 2025



Abstract:To fully leverage the capabilities of diffusion models, we are often interested in optimizing downstream reward functions during inference. While numerous algorithms for reward-guided generation have been recently proposed due to their significance, current approaches predominantly focus on single-shot generation, transitioning from fully noised to denoised states. We propose a novel framework for inference-time reward optimization with diffusion models inspired by evolutionary algorithms. Our approach employs an iterative refinement process consisting of two steps in each iteration: noising and reward-guided denoising. This sequential refinement allows for the gradual correction of errors introduced during reward optimization. Besides, we provide a theoretical guarantee for our framework. Finally, we demonstrate its superior empirical performance in protein and cell-type-specific regulatory DNA design. The code is available at \href{https://github.com/masa-ue/ProDifEvo-Refinement}{https://github.com/masa-ue/ProDifEvo-Refinement}.
Reward-Guided Controlled Generation for Inference-Time Alignment in Diffusion Models: Tutorial and Review
Jan 16, 2025



Abstract:This tutorial provides an in-depth guide on inference-time guidance and alignment methods for optimizing downstream reward functions in diffusion models. While diffusion models are renowned for their generative modeling capabilities, practical applications in fields such as biology often require sample generation that maximizes specific metrics (e.g., stability, affinity in proteins, closeness to target structures). In these scenarios, diffusion models can be adapted not only to generate realistic samples but also to explicitly maximize desired measures at inference time without fine-tuning. This tutorial explores the foundational aspects of such inference-time algorithms. We review these methods from a unified perspective, demonstrating that current techniques -- such as Sequential Monte Carlo (SMC)-based guidance, value-based sampling, and classifier guidance -- aim to approximate soft optimal denoising processes (a.k.a. policies in RL) that combine pre-trained denoising processes with value functions serving as look-ahead functions that predict from intermediate states to terminal rewards. Within this framework, we present several novel algorithms not yet covered in the literature. Furthermore, we discuss (1) fine-tuning methods combined with inference-time techniques, (2) inference-time algorithms based on search algorithms such as Monte Carlo tree search, which have received limited attention in current research, and (3) connections between inference-time algorithms in language models and diffusion models. The code of this tutorial on protein design is available at https://github.com/masa-ue/AlignInversePro
Efficient Fine-Tuning of Single-Cell Foundation Models Enables Zero-Shot Molecular Perturbation Prediction
Dec 18, 2024



Abstract:Predicting transcriptional responses to novel drugs provides a unique opportunity to accelerate biomedical research and advance drug discovery efforts. However, the inherent complexity and high dimensionality of cellular responses, combined with the extremely limited available experimental data, makes the task challenging. In this study, we leverage single-cell foundation models (FMs) pre-trained on tens of millions of single cells, encompassing multiple cell types, states, and disease annotations, to address molecular perturbation prediction. We introduce a drug-conditional adapter that allows efficient fine-tuning by training less than 1% of the original foundation model, thus enabling molecular conditioning while preserving the rich biological representation learned during pre-training. The proposed strategy allows not only the prediction of cellular responses to novel drugs, but also the zero-shot generalization to unseen cell lines. We establish a robust evaluation framework to assess model performance across different generalization tasks, demonstrating state-of-the-art results across all settings, with significant improvements in the few-shot and zero-shot generalization to new cell lines compared to existing baselines.
MolCap-Arena: A Comprehensive Captioning Benchmark on Language-Enhanced Molecular Property Prediction
Nov 01, 2024



Abstract:Bridging biomolecular modeling with natural language information, particularly through large language models (LLMs), has recently emerged as a promising interdisciplinary research area. LLMs, having been trained on large corpora of scientific documents, demonstrate significant potential in understanding and reasoning about biomolecules by providing enriched contextual and domain knowledge. However, the extent to which LLM-driven insights can improve performance on complex predictive tasks (e.g., toxicity) remains unclear. Further, the extent to which relevant knowledge can be extracted from LLMs also remains unknown. In this study, we present Molecule Caption Arena: the first comprehensive benchmark of LLM-augmented molecular property prediction. We evaluate over twenty LLMs, including both general-purpose and domain-specific molecule captioners, across diverse prediction tasks. To this goal, we introduce a novel, battle-based rating system. Our findings confirm the ability of LLM-extracted knowledge to enhance state-of-the-art molecular representations, with notable model-, prompt-, and dataset-specific variations. Code, resources, and data are available at github.com/Genentech/molcap-arena.
Fine-Tuning Discrete Diffusion Models via Reward Optimization with Applications to DNA and Protein Design
Oct 17, 2024



Abstract:Recent studies have demonstrated the strong empirical performance of diffusion models on discrete sequences across domains from natural language to biological sequence generation. For example, in the protein inverse folding task, conditional diffusion models have achieved impressive results in generating natural-like sequences that fold back into the original structure. However, practical design tasks often require not only modeling a conditional distribution but also optimizing specific task objectives. For instance, we may prefer protein sequences with high stability. To address this, we consider the scenario where we have pre-trained discrete diffusion models that can generate natural-like sequences, as well as reward models that map sequences to task objectives. We then formulate the reward maximization problem within discrete diffusion models, analogous to reinforcement learning (RL), while minimizing the KL divergence against pretrained diffusion models to preserve naturalness. To solve this RL problem, we propose a novel algorithm, DRAKES, that enables direct backpropagation of rewards through entire trajectories generated by diffusion models, by making the originally non-differentiable trajectories differentiable using the Gumbel-Softmax trick. Our theoretical analysis indicates that our approach can generate sequences that are both natural-like and yield high rewards. While similar tasks have been recently explored in diffusion models for continuous domains, our work addresses unique algorithmic and theoretical challenges specific to discrete diffusion models, which arise from their foundation in continuous-time Markov chains rather than Brownian motion. Finally, we demonstrate the effectiveness of DRAKES in generating DNA and protein sequences that optimize enhancer activity and protein stability, respectively, important tasks for gene therapies and protein-based therapeutics.
A mechanistically interpretable neural network for regulatory genomics
Oct 08, 2024



Abstract:Deep neural networks excel in mapping genomic DNA sequences to associated readouts (e.g., protein-DNA binding). Beyond prediction, the goal of these networks is to reveal to scientists the underlying motifs (and their syntax) which drive genome regulation. Traditional methods that extract motifs from convolutional filters suffer from the uninterpretable dispersion of information across filters and layers. Other methods which rely on importance scores can be unstable and unreliable. Instead, we designed a novel mechanistically interpretable architecture for regulatory genomics, where motifs and their syntax are directly encoded and readable from the learned weights and activations. We provide theoretical and empirical evidence of our architecture's full expressivity, while still being highly interpretable. Through several experiments, we show that our architecture excels in de novo motif discovery and motif instance calling, is robust to variable sequence contexts, and enables fully interpretable generation of novel functional sequences.
Derivative-Free Guidance in Continuous and Discrete Diffusion Models with Soft Value-Based Decoding
Aug 15, 2024



Abstract:Diffusion models excel at capturing the natural design spaces of images, molecules, DNA, RNA, and protein sequences. However, rather than merely generating designs that are natural, we often aim to optimize downstream reward functions while preserving the naturalness of these design spaces. Existing methods for achieving this goal often require ``differentiable'' proxy models (\textit{e.g.}, classifier guidance or DPS) or involve computationally expensive fine-tuning of diffusion models (\textit{e.g.}, classifier-free guidance, RL-based fine-tuning). In our work, we propose a new method to address these challenges. Our algorithm is an iterative sampling method that integrates soft value functions, which looks ahead to how intermediate noisy states lead to high rewards in the future, into the standard inference procedure of pre-trained diffusion models. Notably, our approach avoids fine-tuning generative models and eliminates the need to construct differentiable models. This enables us to (1) directly utilize non-differentiable features/reward feedback, commonly used in many scientific domains, and (2) apply our method to recent discrete diffusion models in a principled way. Finally, we demonstrate the effectiveness of our algorithm across several domains, including image generation, molecule generation, and DNA/RNA sequence generation. The code is available at \href{https://github.com/masa-ue/SVDD}{https://github.com/masa-ue/SVDD}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge