Thomas M. Sutter
You Only Train Once: Differentiable Subset Selection for Omics Data
Dec 19, 2025



Abstract:Selecting compact and informative gene subsets from single-cell transcriptomic data is essential for biomarker discovery, improving interpretability, and cost-effective profiling. However, most existing feature selection approaches either operate as multi-stage pipelines or rely on post hoc feature attribution, making selection and prediction weakly coupled. In this work, we present YOTO (you only train once), an end-to-end framework that jointly identifies discrete gene subsets and performs prediction within a single differentiable architecture. In our model, the prediction task directly guides which genes are selected, while the learned subsets, in turn, shape the predictive representation. This closed feedback loop enables the model to iteratively refine both what it selects and how it predicts during training. Unlike existing approaches, YOTO enforces sparsity so that only the selected genes contribute to inference, eliminating the need to train additional downstream classifiers. Through a multi-task learning design, the model learns shared representations across related objectives, allowing partially labeled datasets to inform one another, and discovering gene subsets that generalize across tasks without additional training steps. We evaluate YOTO on two representative single-cell RNA-seq datasets, showing that it consistently outperforms state-of-the-art baselines. These results demonstrate that sparse, end-to-end, multi-task gene subset selection improves predictive performance and yields compact and meaningful gene subsets, advancing biomarker discovery and single-cell analysis.
Enhancing Radiology Report Generation and Visual Grounding using Reinforcement Learning
Dec 11, 2025Abstract:Recent advances in vision-language models (VLMs) have improved Chest X-ray (CXR) interpretation in multiple aspects. However, many medical VLMs rely solely on supervised fine-tuning (SFT), which optimizes next-token prediction without evaluating answer quality. In contrast, reinforcement learning (RL) can incorporate task-specific feedback, and its combination with explicit intermediate reasoning ("thinking") has demonstrated substantial gains on verifiable math and coding tasks. To investigate the effects of RL and thinking in a CXR VLM, we perform large-scale SFT on CXR data to build an updated RadVLM based on Qwen3-VL, followed by a cold-start SFT stage that equips the model with basic thinking ability. We then apply Group Relative Policy Optimization (GRPO) with clinically grounded, task-specific rewards for report generation and visual grounding, and run matched RL experiments on both domain-specific and general-domain Qwen3-VL variants, with and without thinking. Across these settings, we find that while strong SFT remains crucial for high base performance, RL provides additional gains on both tasks, whereas explicit thinking does not appear to further improve results. Under a unified evaluation pipeline, the RL-optimized RadVLM models outperform their baseline counterparts and reach state-of-the-art performance on both report generation and grounding, highlighting clinically aligned RL as a powerful complement to SFT for medical VLMs.
RadVLM: A Multitask Conversational Vision-Language Model for Radiology
Feb 05, 2025



Abstract:The widespread use of chest X-rays (CXRs), coupled with a shortage of radiologists, has driven growing interest in automated CXR analysis and AI-assisted reporting. While existing vision-language models (VLMs) show promise in specific tasks such as report generation or abnormality detection, they often lack support for interactive diagnostic capabilities. In this work we present RadVLM, a compact, multitask conversational foundation model designed for CXR interpretation. To this end, we curate a large-scale instruction dataset comprising over 1 million image-instruction pairs containing both single-turn tasks -- such as report generation, abnormality classification, and visual grounding -- and multi-turn, multi-task conversational interactions. After fine-tuning RadVLM on this instruction dataset, we evaluate it across different tasks along with re-implemented baseline VLMs. Our results show that RadVLM achieves state-of-the-art performance in conversational capabilities and visual grounding while remaining competitive in other radiology tasks. Ablation studies further highlight the benefit of joint training across multiple tasks, particularly for scenarios with limited annotated data. Together, these findings highlight the potential of RadVLM as a clinically relevant AI assistant, providing structured CXR interpretation and conversational capabilities to support more effective and accessible diagnostic workflows.
Weakly-Supervised Multimodal Learning on MIMIC-CXR
Nov 15, 2024



Abstract:Multimodal data integration and label scarcity pose significant challenges for machine learning in medical settings. To address these issues, we conduct an in-depth evaluation of the newly proposed Multimodal Variational Mixture-of-Experts (MMVM) VAE on the challenging MIMIC-CXR dataset. Our analysis demonstrates that the MMVM VAE consistently outperforms other multimodal VAEs and fully supervised approaches, highlighting its strong potential for real-world medical applications.
Anomaly Detection by Context Contrasting
May 29, 2024



Abstract:Anomaly Detection focuses on identifying samples that deviate from the norm. When working with high-dimensional data such as images, a crucial requirement for detecting anomalous patterns is learning lower-dimensional representations that capture normal concepts seen during training. Recent advances in self-supervised learning have shown great promise in this regard. However, many of the most successful self-supervised anomaly detection methods assume prior knowledge about the structure of anomalies and leverage synthetic anomalies during training. Yet, in many real-world applications, we do not know what to expect from unseen data, and we can solely leverage knowledge about normal data. In this work, we propose Con2, which addresses this problem by setting normal training data into distinct contexts while preserving its normal properties, letting us observe the data from different perspectives. Unseen normal data consequently adheres to learned context representations while anomalies fail to do so, letting us detect them without any knowledge about anomalies during training. Our experiments demonstrate that our approach achieves state-of-the-art performance on various benchmarks while exhibiting superior performance in a more realistic healthcare setting, where knowledge about potential anomalies is often scarce.
Unity by Diversity: Improved Representation Learning in Multimodal VAEs
Mar 08, 2024



Abstract:Variational Autoencoders for multimodal data hold promise for many tasks in data analysis, such as representation learning, conditional generation, and imputation. Current architectures either share the encoder output, decoder input, or both across modalities to learn a shared representation. Such architectures impose hard constraints on the model. In this work, we show that a better latent representation can be obtained by replacing these hard constraints with a soft constraint. We propose a new mixture-of-experts prior, softly guiding each modality's latent representation towards a shared aggregate posterior. This approach results in a superior latent representation and allows each encoding to preserve information from its uncompressed original features better. In extensive experiments on multiple benchmark datasets and a challenging real-world neuroscience data set, we show improved learned latent representations and imputation of missing data modalities compared to existing methods.
M-mode Based Prediction of Ejection Fraction using Echocardiograms
Sep 07, 2023



Abstract:Early detection of cardiac dysfunction through routine screening is vital for diagnosing cardiovascular diseases. An important metric of cardiac function is the left ventricular ejection fraction (EF), where lower EF is associated with cardiomyopathy. Echocardiography is a popular diagnostic tool in cardiology, with ultrasound being a low-cost, real-time, and non-ionizing technology. However, human assessment of echocardiograms for calculating EF is time-consuming and expertise-demanding, raising the need for an automated approach. In this work, we propose using the M(otion)-mode of echocardiograms for estimating the EF and classifying cardiomyopathy. We generate multiple artificial M-mode images from a single echocardiogram and combine them using off-the-shelf model architectures. Additionally, we extend contrastive learning (CL) to cardiac imaging to learn meaningful representations from exploiting structures in unlabeled data allowing the model to achieve high accuracy, even with limited annotations. Our experiments show that the supervised setting converges with only ten modes and is comparable to the baseline method while bypassing its cumbersome training process and being computationally much more efficient. Furthermore, CL using M-mode images is helpful for limited data scenarios, such as having labels for only 200 patients, which is common in medical applications.
Differentiable Random Partition Models
May 26, 2023



Abstract:Partitioning a set of elements into an unknown number of mutually exclusive subsets is essential in many machine learning problems. However, assigning elements, such as samples in a dataset or neurons in a network layer, to an unknown and discrete number of subsets is inherently non-differentiable, prohibiting end-to-end gradient-based optimization of parameters. We overcome this limitation by proposing a novel two-step method for inferring partitions, which allows its usage in variational inference tasks. This new approach enables reparameterized gradients with respect to the parameters of the new random partition model. Our method works by inferring the number of elements per subset and, second, by filling these subsets in a learned order. We highlight the versatility of our general-purpose approach on three different challenging experiments: variational clustering, inference of shared and independent generative factors under weak supervision, and multitask learning.
Continuous Relaxation For The Multivariate Non-Central Hypergeometric Distribution
Mar 03, 2022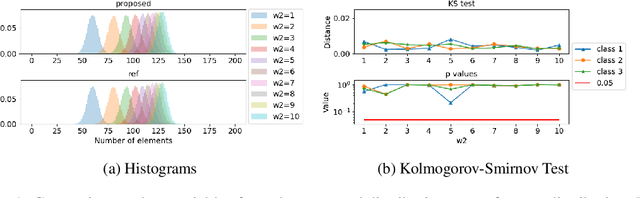
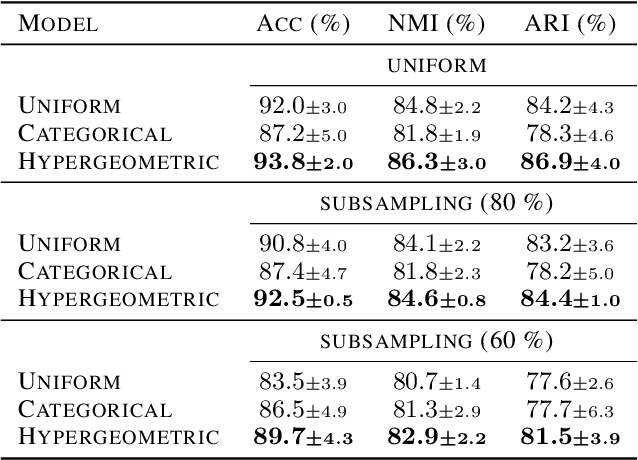
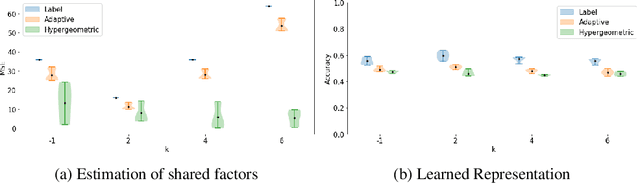
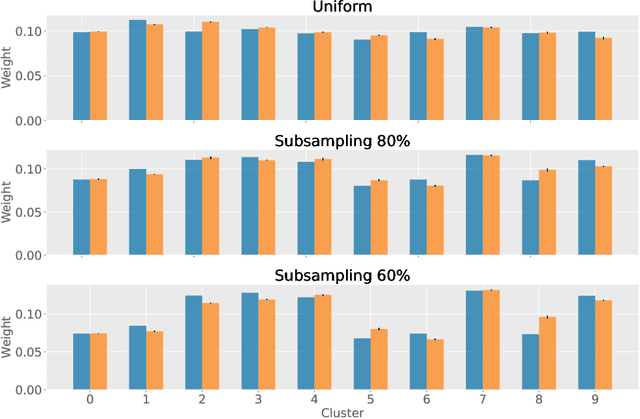
Abstract:Partitioning a set of elements into a given number of groups of a priori unknown sizes is an important task in many applications. Due to hard constraints, it is a non-differentiable problem which prohibits its direct use in modern machine learning frameworks. Hence, previous works mostly fall back on suboptimal heuristics or simplified assumptions. The multivariate hypergeometric distribution offers a probabilistic formulation of how to distribute a given number of samples across multiple groups. Unfortunately, as a discrete probability distribution, it neither is differentiable. In this work, we propose a continuous relaxation for the multivariate non-central hypergeometric distribution. We introduce an efficient and numerically stable sampling procedure. This enables reparameterized gradients for the hypergeometric distribution and its integration into automatic differentiation frameworks. We highlight the applicability and usability of the proposed formulation on two different common machine learning tasks.
On the Limitations of Multimodal VAEs
Oct 08, 2021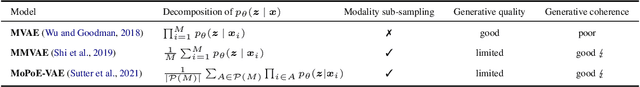
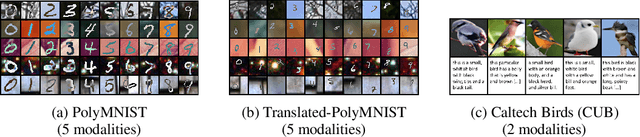
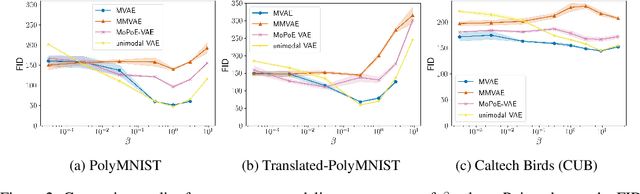
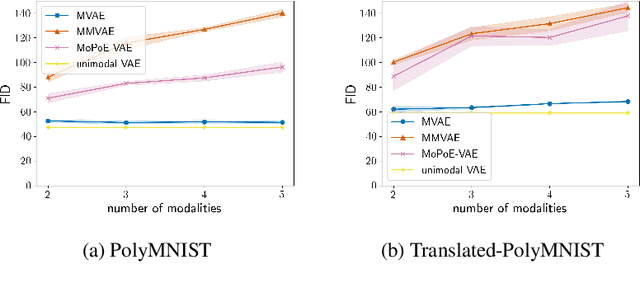
Abstract:Multimodal variational autoencoders (VAEs) have shown promise as efficient generative models for weakly-supervised data. Yet, despite their advantage of weak supervision, they exhibit a gap in generative quality compared to unimodal VAEs, which are completely unsupervised. In an attempt to explain this gap, we uncover a fundamental limitation that applies to a large family of mixture-based multimodal VAEs. We prove that the sub-sampling of modalities enforces an undesirable upper bound on the multimodal ELBO and thereby limits the generative quality of the respective models. Empirically, we showcase the generative quality gap on both synthetic and real data and present the tradeoffs between different variants of multimodal VAEs. We find that none of the existing approaches fulfills all desired criteria of an effective multimodal generative model when applied on more complex datasets than those used in previous benchmarks. In summary, we identify, formalize, and validate fundamental limitations of VAE-based approaches for modeling weakly-supervised data and discuss implications for real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge