Emanuele Palumbo
Hybrid Modeling of Photoplethysmography for Non-invasive Monitoring of Cardiovascular Parameters
Nov 18, 2025Abstract:Continuous cardiovascular monitoring can play a key role in precision health. However, some fundamental cardiac biomarkers of interest, including stroke volume and cardiac output, require invasive measurements, e.g., arterial pressure waveforms (APW). As a non-invasive alternative, photoplethysmography (PPG) measurements are routinely collected in hospital settings. Unfortunately, the prediction of key cardiac biomarkers from PPG instead of APW remains an open challenge, further complicated by the scarcity of annotated PPG measurements. As a solution, we propose a hybrid approach that uses hemodynamic simulations and unlabeled clinical data to estimate cardiovascular biomarkers directly from PPG signals. Our hybrid model combines a conditional variational autoencoder trained on paired PPG-APW data with a conditional density estimator of cardiac biomarkers trained on labeled simulated APW segments. As a key result, our experiments demonstrate that the proposed approach can detect fluctuations of cardiac output and stroke volume and outperform a supervised baseline in monitoring temporal changes in these biomarkers.
From Logits to Hierarchies: Hierarchical Clustering made Simple
Oct 10, 2024



Abstract:The structure of many real-world datasets is intrinsically hierarchical, making the modeling of such hierarchies a critical objective in both unsupervised and supervised machine learning. Recently, novel approaches for hierarchical clustering with deep architectures have been proposed. In this work, we take a critical perspective on this line of research and demonstrate that many approaches exhibit major limitations when applied to realistic datasets, partly due to their high computational complexity. In particular, we show that a lightweight procedure implemented on top of pre-trained non-hierarchical clustering models outperforms models designed specifically for hierarchical clustering. Our proposed approach is computationally efficient and applicable to any pre-trained clustering model that outputs logits, without requiring any fine-tuning. To highlight the generality of our findings, we illustrate how our method can also be applied in a supervised setup, recovering meaningful hierarchies from a pre-trained ImageNet classifier.
Identifiability Results for Multimodal Contrastive Learning
Mar 16, 2023



Abstract:Contrastive learning is a cornerstone underlying recent progress in multi-view and multimodal learning, e.g., in representation learning with image/caption pairs. While its effectiveness is not yet fully understood, a line of recent work reveals that contrastive learning can invert the data generating process and recover ground truth latent factors shared between views. In this work, we present new identifiability results for multimodal contrastive learning, showing that it is possible to recover shared factors in a more general setup than the multi-view setting studied previously. Specifically, we distinguish between the multi-view setting with one generative mechanism (e.g., multiple cameras of the same type) and the multimodal setting that is characterized by distinct mechanisms (e.g., cameras and microphones). Our work generalizes previous identifiability results by redefining the generative process in terms of distinct mechanisms with modality-specific latent variables. We prove that contrastive learning can block-identify latent factors shared between modalities, even when there are nontrivial dependencies between factors. We empirically verify our identifiability results with numerical simulations and corroborate our findings on a complex multimodal dataset of image/text pairs. Zooming out, our work provides a theoretical basis for multimodal representation learning and explains in which settings multimodal contrastive learning can be effective in practice.
On the Limitations of Multimodal VAEs
Oct 08, 2021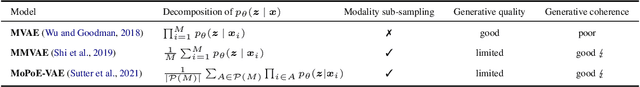
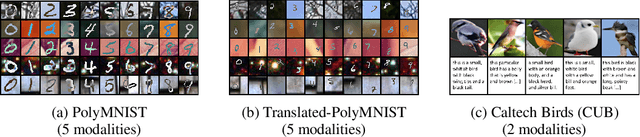
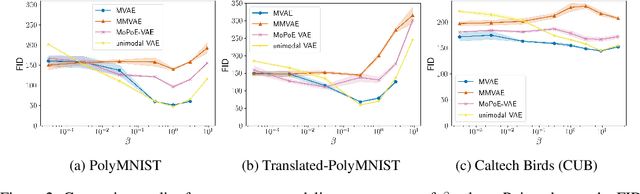
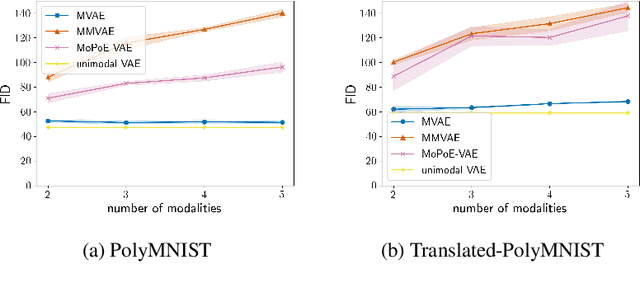
Abstract:Multimodal variational autoencoders (VAEs) have shown promise as efficient generative models for weakly-supervised data. Yet, despite their advantage of weak supervision, they exhibit a gap in generative quality compared to unimodal VAEs, which are completely unsupervised. In an attempt to explain this gap, we uncover a fundamental limitation that applies to a large family of mixture-based multimodal VAEs. We prove that the sub-sampling of modalities enforces an undesirable upper bound on the multimodal ELBO and thereby limits the generative quality of the respective models. Empirically, we showcase the generative quality gap on both synthetic and real data and present the tradeoffs between different variants of multimodal VAEs. We find that none of the existing approaches fulfills all desired criteria of an effective multimodal generative model when applied on more complex datasets than those used in previous benchmarks. In summary, we identify, formalize, and validate fundamental limitations of VAE-based approaches for modeling weakly-supervised data and discuss implications for real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge