Daphné Chopard
You Only Train Once: Differentiable Subset Selection for Omics Data
Dec 19, 2025



Abstract:Selecting compact and informative gene subsets from single-cell transcriptomic data is essential for biomarker discovery, improving interpretability, and cost-effective profiling. However, most existing feature selection approaches either operate as multi-stage pipelines or rely on post hoc feature attribution, making selection and prediction weakly coupled. In this work, we present YOTO (you only train once), an end-to-end framework that jointly identifies discrete gene subsets and performs prediction within a single differentiable architecture. In our model, the prediction task directly guides which genes are selected, while the learned subsets, in turn, shape the predictive representation. This closed feedback loop enables the model to iteratively refine both what it selects and how it predicts during training. Unlike existing approaches, YOTO enforces sparsity so that only the selected genes contribute to inference, eliminating the need to train additional downstream classifiers. Through a multi-task learning design, the model learns shared representations across related objectives, allowing partially labeled datasets to inform one another, and discovering gene subsets that generalize across tasks without additional training steps. We evaluate YOTO on two representative single-cell RNA-seq datasets, showing that it consistently outperforms state-of-the-art baselines. These results demonstrate that sparse, end-to-end, multi-task gene subset selection improves predictive performance and yields compact and meaningful gene subsets, advancing biomarker discovery and single-cell analysis.
Towards Foundation Models for Critical Care Time Series
Nov 25, 2024



Abstract:Notable progress has been made in generalist medical large language models across various healthcare areas. However, large-scale modeling of in-hospital time series data - such as vital signs, lab results, and treatments in critical care - remains underexplored. Existing datasets are relatively small, but combining them can enhance patient diversity and improve model robustness. To effectively utilize these combined datasets for large-scale modeling, it is essential to address the distribution shifts caused by varying treatment policies, necessitating the harmonization of treatment variables across the different datasets. This work aims to establish a foundation for training large-scale multi-variate time series models on critical care data and to provide a benchmark for machine learning models in transfer learning across hospitals to study and address distribution shift challenges. We introduce a harmonized dataset for sequence modeling and transfer learning research, representing the first large-scale collection to include core treatment variables. Future plans involve expanding this dataset to support further advancements in transfer learning and the development of scalable, generalizable models for critical healthcare applications.
Weakly-Supervised Multimodal Learning on MIMIC-CXR
Nov 15, 2024



Abstract:Multimodal data integration and label scarcity pose significant challenges for machine learning in medical settings. To address these issues, we conduct an in-depth evaluation of the newly proposed Multimodal Variational Mixture-of-Experts (MMVM) VAE on the challenging MIMIC-CXR dataset. Our analysis demonstrates that the MMVM VAE consistently outperforms other multimodal VAEs and fully supervised approaches, highlighting its strong potential for real-world medical applications.
Automatic Classification of General Movements in Newborns
Nov 14, 2024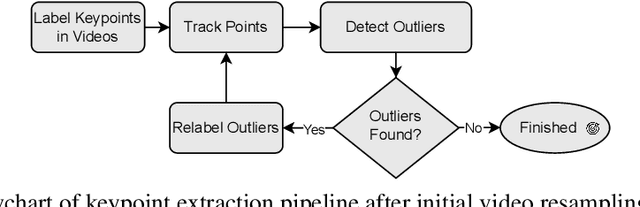
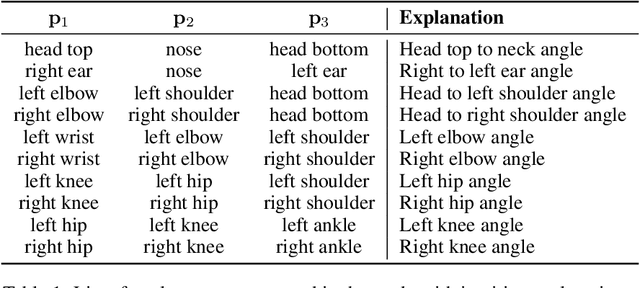

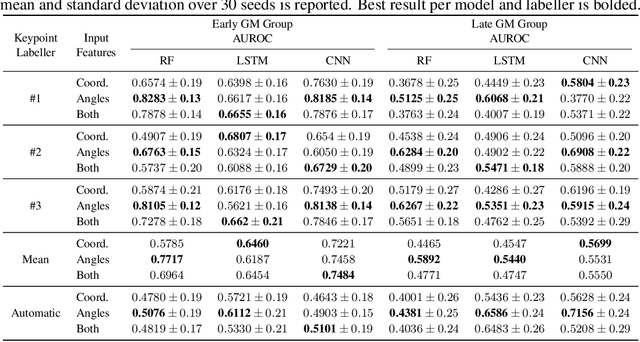
Abstract:General movements (GMs) are spontaneous, coordinated body movements in infants that offer valuable insights into the developing nervous system. Assessed through the Prechtl GM Assessment (GMA), GMs are reliable predictors for neurodevelopmental disorders. However, GMA requires specifically trained clinicians, who are limited in number. To scale up newborn screening, there is a need for an algorithm that can automatically classify GMs from infant video recordings. This data poses challenges, including variability in recording length, device type, and setting, with each video coarsely annotated for overall movement quality. In this work, we introduce a tool for extracting features from these recordings and explore various machine learning techniques for automated GM classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge